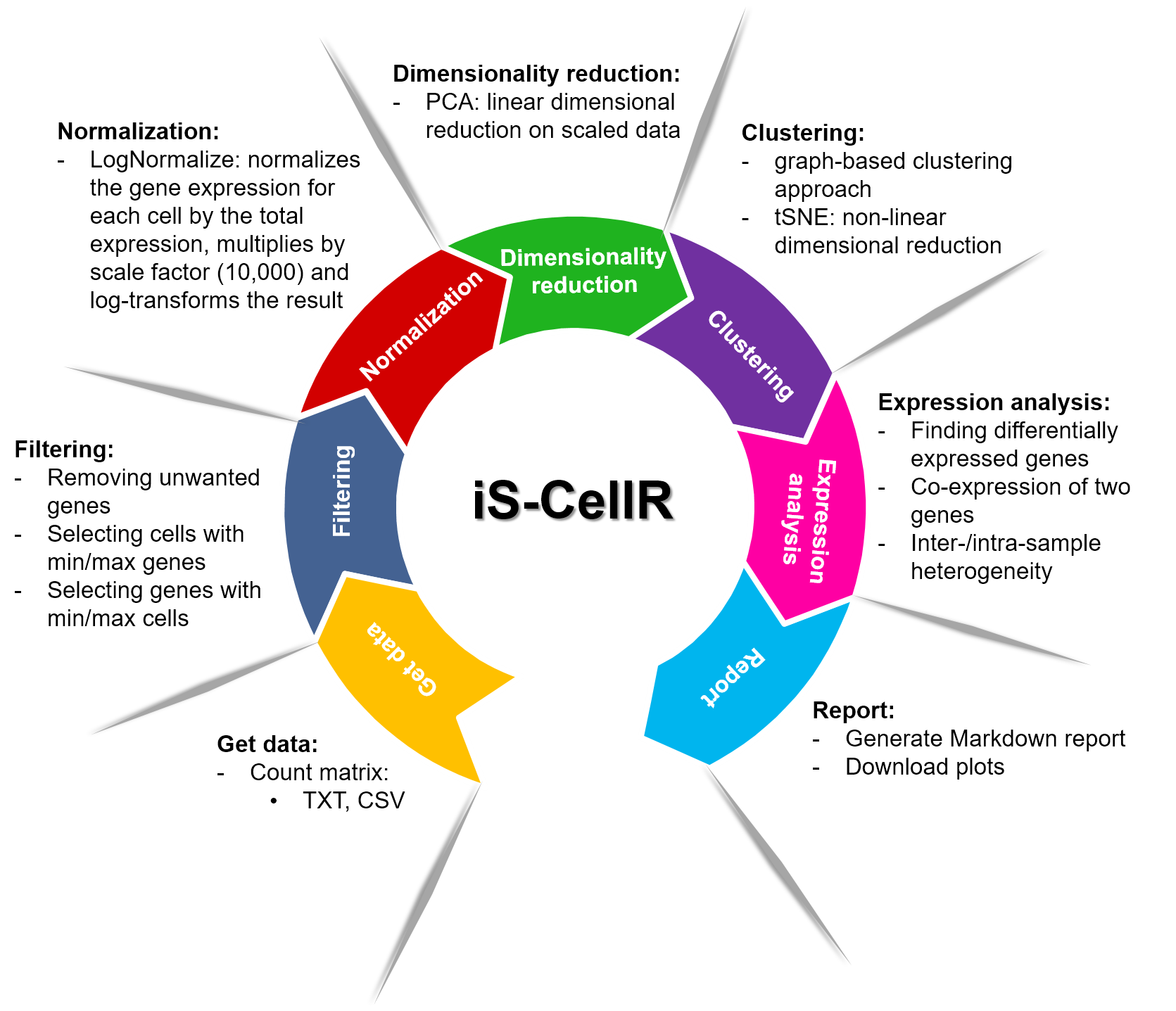
iS-CellR (Interactive platform for Single-cell RNAseq) is a web-based Shiny application designed to provide a comprehensive analysis of single-cell RNA sequencing data. iS-CellR provides a fast method for filtering and normalization of raw data, dimensionality reductions (linear and non-linear) to identify cell types clusters, differential gene expression analysis to locate markers, and inter-/intra-sample heterogeneity analysis. iS-CellR integrates the Seurat package with Shiny's reactive programming framework and interactive visualization using plotly library. iS-CellR runs on any modern web browser and provides access to powerful R libraries through a graphical user interface. Each session of iS-CellR allows the user to share, reproduce and archive results without requiring programming skills.
iS-CellR pipeline overview is illustrated in the figure. iS-CellR is organized into a seven-step process for complete scRNA-seq analysis.
1). Install Docker (v >= 18.02.0-ce)
Download and install Docker from https://docs.docker.com/install/
2). Install R (v >= 3.2)
Download and install R from http://cran.us.r-project.org/
3). Install R studio (Optional)
Download and install RStudio Desktop from http://rstudio.org/download/desktop
4). Install Shiny package
install.packages(“shiny”) # inside R console
install.packages("devtools")
devtools::install_github("rstudio/shiny", dependencies=FALSE)
runUrl('https://github.com/immcore/iS-CellR/archive/master.zip')
# or
shiny::runGitHub("iS-CellR", "immcore")
# or
runApp(/fullpath/iS-CellR) # provide full path to iS-CellR folder
To run container use the command below:
docker run --rm -p 3838:3838 immcore/is-cellr After that check with your browser at addresses plus the port 3838 : http://0.0.0.0:3838/
1). Seurat installaton
When you see the following warning message in the R console:
Warning: Installed Rcpp (0.12.12) different from Rcpp used to build dplyr (0.12.11).
Please reinstall dplyr to avoid random crashes or undefined behavior.
install.packages("dplyr", type = "source")
library(dplyr)
If you are using older R versions, installing from source causes errors on some standard Mac installs due to R being compiled using gfortran-4.8.
The error will look something like:
-L/usr/local/lib/gcc/x86_64-apple-darwin13.0.0/4.8.2'
ld: library not found for -lgfortran
To fix the above error type following commands in terminal. * sudo permissions required*
curl -O http://r.research.att.com/libs/gfortran-4.8.2-darwin13.tar.bz2
sudo tar fvxz gfortran-4.8.2-darwin13.tar.bz2 -C /
It is important that the input file should follow the same format as descibed below.
To get started, please load in a CSV/TSV separated values file. The file should contain the count data where:
- Columns are Cells
- Rows are Genes
- Optional: Cell names or Cell type in Column header followed by Cell id. eg. Tcell_TC1_fcount_1.
iS-CellR v1.1 now supports following inputs:
- 10X genomics Cellranger output
- Precomputed Rds files
There is one matrix file for Malignant dataset from Tirosh et al., 2016 (Main file for the analysis) and genes file (For STEP: Discriminating marker genes).