The International Brain Laboratory Viewer is a simple and fast 3D interactive visualization tool based on VTK that uses GPU accelerated volume and surface rendering. It runs on python 3.8+ and it can be embed in Jupyter Lab/Notebook and Qt user interfaces.
This viewer is featured with an optional QT user interface with more advanced functionalities like dynamic statistics. In the terminal, type iblviewer and see what parameters are available. The viewer launches by default with a Qt UI.
Most of the viewer makes VTK usable as an interactive application and you may use it as such. Just use from iblviewer.application import Viewer
The small added part related to IBL allows scientists to view their data and models for further analysis. From electrophysiological data to neuronal connectivity, this tool allows simple and effective 3D visualization for many use-cases like multi-slicing and time series even on volumes. In that case, you will use from iblviewer.mouse_brain import MouseBrainViewer
pip install iblviewerIf you wish to use this viewer with International Brain Laboratory data sets and libraries, you will need ibllib:
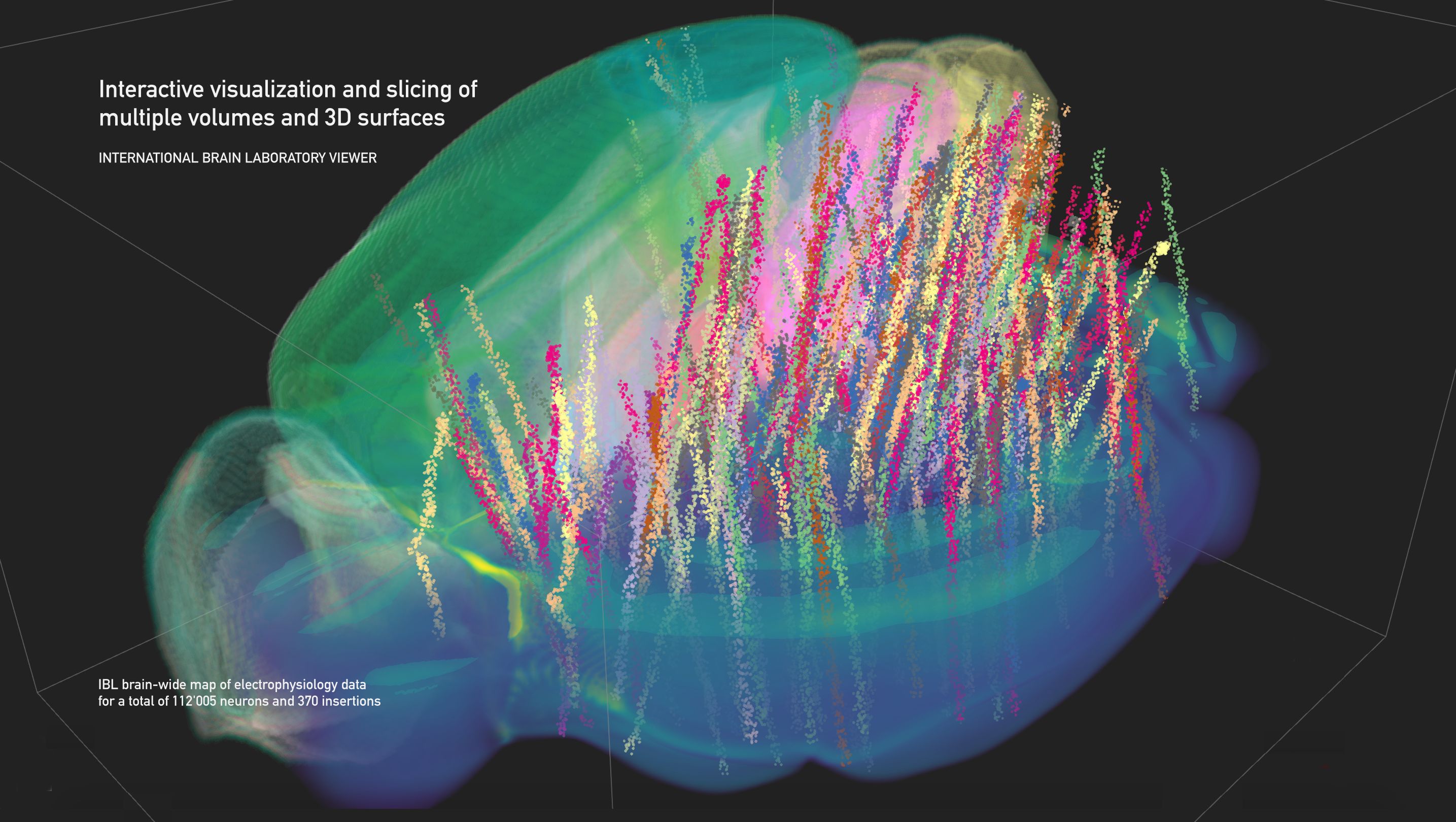
pip install ibllibAn example of mouse brain-wide map of electrophysiological recordings (seen here as point neurons) in the Allen Brain Atlas CCF v3 with both DWI and segmented volumes.
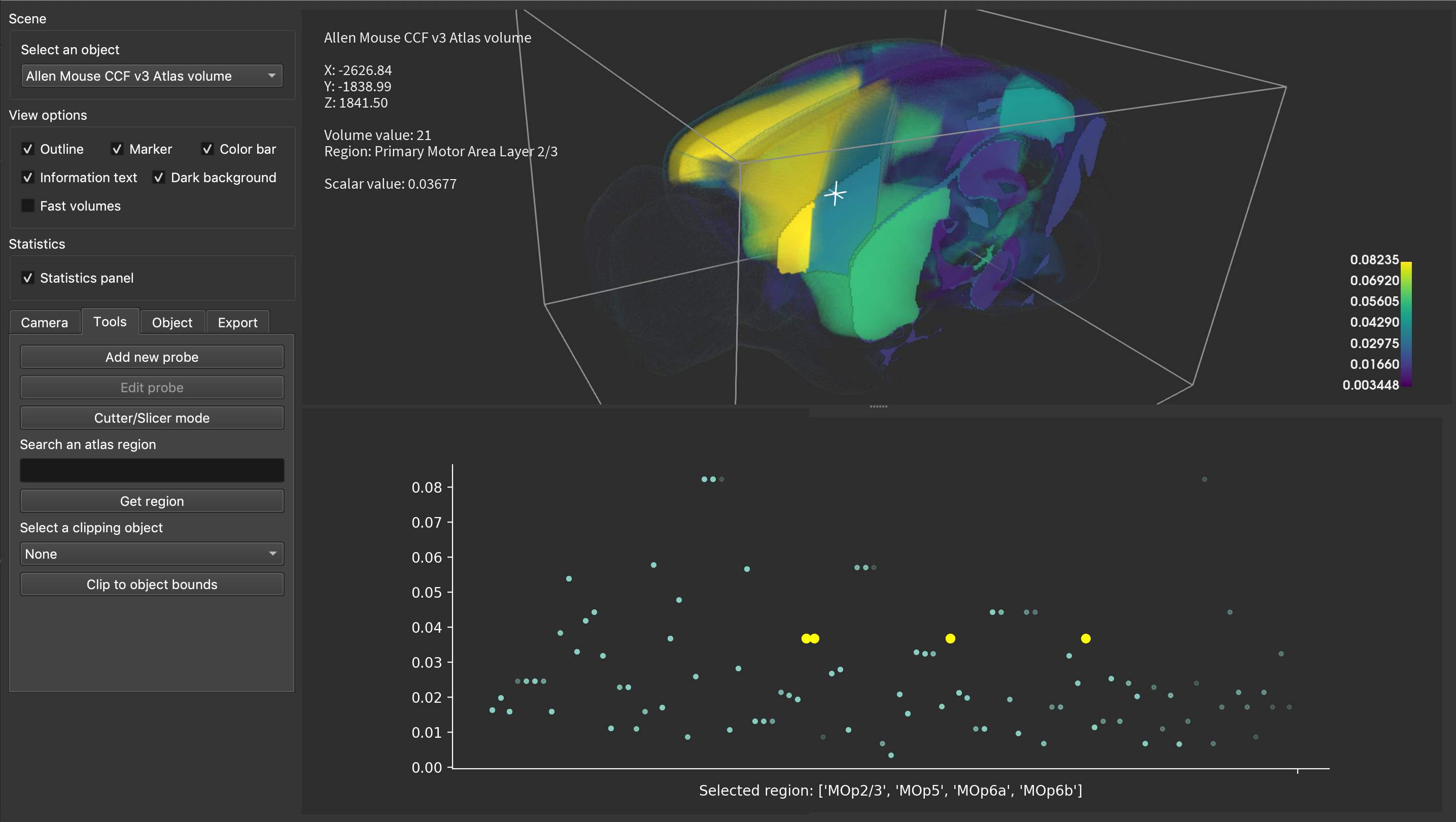
Another example of the Qt UI with additional features like custom interactive statistics, in this case with matplotlib.
If at some point it complains about failing to uninstall vtk, it's likely that vtk is already installed within your conda environment and that it causes troubles (even if it's the proper version). Run the following:
conda uninstall vtk
pip install vtkThis will uninstall vtk and reinstall it (version 9+) with pip.
If you have installed IBLViewer (see below) and you to update to the latest version, run:
pip install -U iblviewerIn rare cases like observed on Windows once, updating fails and the user doesn't know about it. Reinstall iblviewer:
pip uninstall iblviewer
pip install iblviewerWrite iblviewer in the command line to start the viewer, add --help for info about arguments.
You may launch examples/demos from the command line too, write iblviewer, hit TAB key twice and a list of names are given, like iblviewer-volume-mapping-demo. There's a demo for headless rendering when you only need to execute code and produce an image or video.
If you wish to run your own code, here are steps below.
Code to run the launcher with arguments from the command line (such as using the Qt UI)
from iblviewer.launcher import IBLViewer
viewer = IBLViewer()
viewer.launch()If you're not interested in the Qt UI, you may either directly use the VTK viewer below or the neuroscience one.
Sample code to run the generic VTK viewer:
from iblviewer.application import Viewer
viewer = Viewer()
viewer.initialize(embed_ui=True)
# Add some random point data
points = np.random.random((500, 3)) * 1000
viewer.add_points(points)
# Select and autofocus the last added object (the points)
viewer.select(-1)
# viewer.select(points) or viewer.select(points.name) yield the same result
viewer.show()Sample code to run the mouse atlas viewer:
from iblviewer.mouse_brain import MouseBrainViewer
viewer = MouseBrainViewer()
# See initialize parameters for more choices
viewer.initialize(resolution=50, mapping='Allen', add_atlas=True,
add_dwi=False, embed_ui=True, jupyter=False)
viewer.show()Volumetric time series of values assigned to brain regions.
Point neurons and connectivity data.
Insertion probes, or how to display lines made of an heterogeneous amount of points. This example requires valid credentials to IBL back-end.
Since this tool is built on top of VTK and vedo, a wrapper for VTK that makes it easy to use, you have endless possibilities for plotting and visualizing your data.
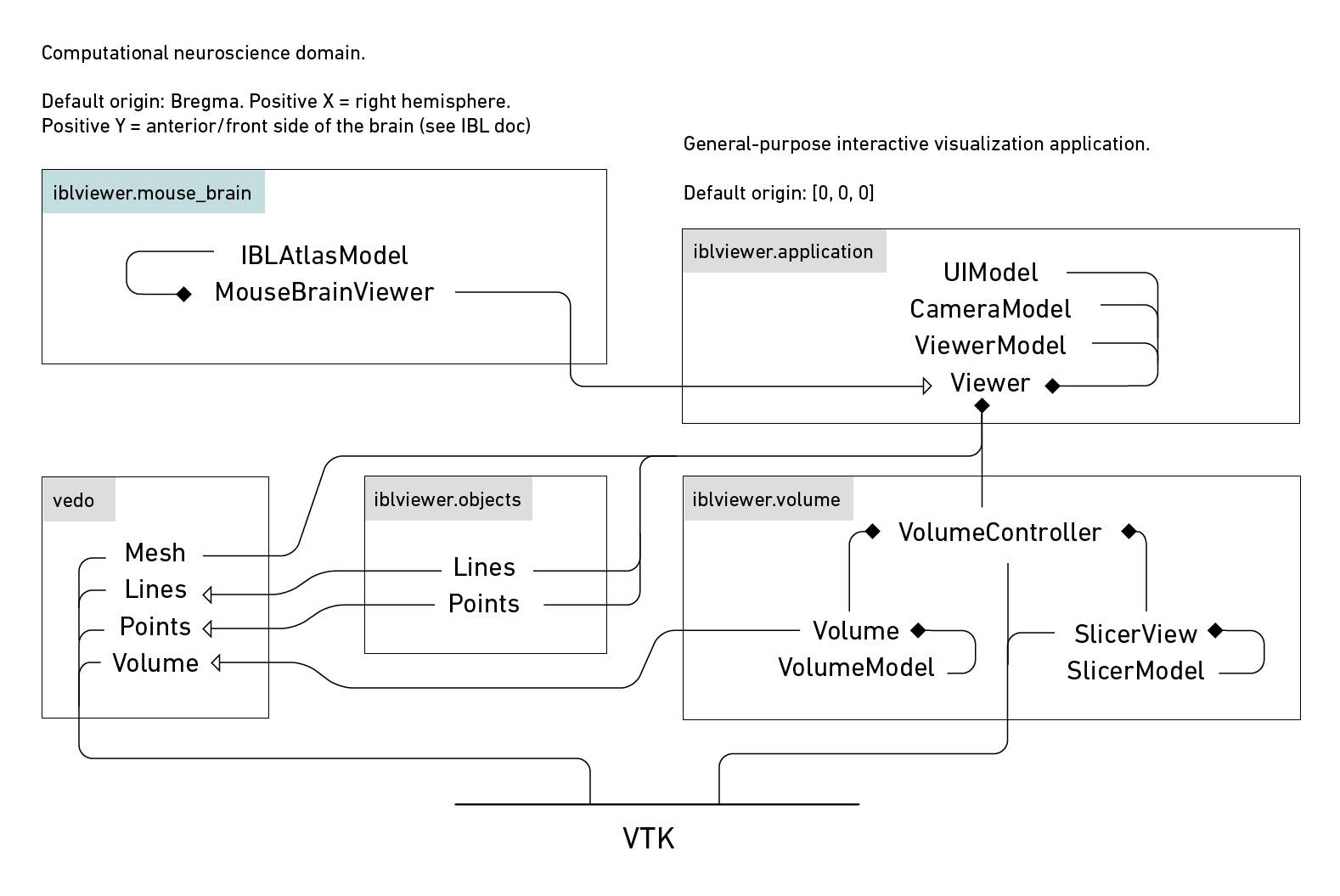
This application relies on the well-known pattern MVC (Model-View-Controller). By decoupling elements, it is easy to extend the application and to customize it for your needs.
This application partly relies on vedo, a wrapper for vtk python (that makes it easier to use). When it comes to volume rendering, vedo and its challenger pyvista are lacking. When you start working on scientific analysis and modeling using volumetric data (combined with surface meshes if you wish), this viewer comes in handy.
vedo and pyvista, two packages doing the same thing really, that is wrapping vtk python in an easily accessible way are great to start with. If you want to build an application with optimized updating mechanisms (that are part of VTK already), vedo and pyvista are not made for this per say. So we keep here the useful parts of vedo and for all the rest, we use vtk python.
IBLViewer adds the following features:
- simple but powerful features
- update-oriented rather than destroy-create
- per-context UI and state
- slicer that can be controlled by the UI or by code
- interactive volumetric data mapping
- mixing volumes and surfaces
Feel free to request features, submit PRs and raise issues.
Nicolas Antille, International Brain Laboratory, 2021
Thanks Marco Musy and Federico Claudi for their support in using vedo. Check out the tool that Federico made, called brainrender, a tool that leverages surface rendering to create great scientific figures.
From International Brain Laboratory: Thanks professor Alexandre Pouget, Berk Gerçek, Guido Meijer, Leenoy Meshulam and Alessandro Santos for their constructive feedbacks and guidance. Thanks Olivier Winter, Gaelle Chapuis and Shan Shen for their support.
The project was initiated and funded by the laboratory of professor Alexandre Pouget, University of Geneva, Faculty of Medecine, Basic Neuroscience which participates to International Brain Laboratory.