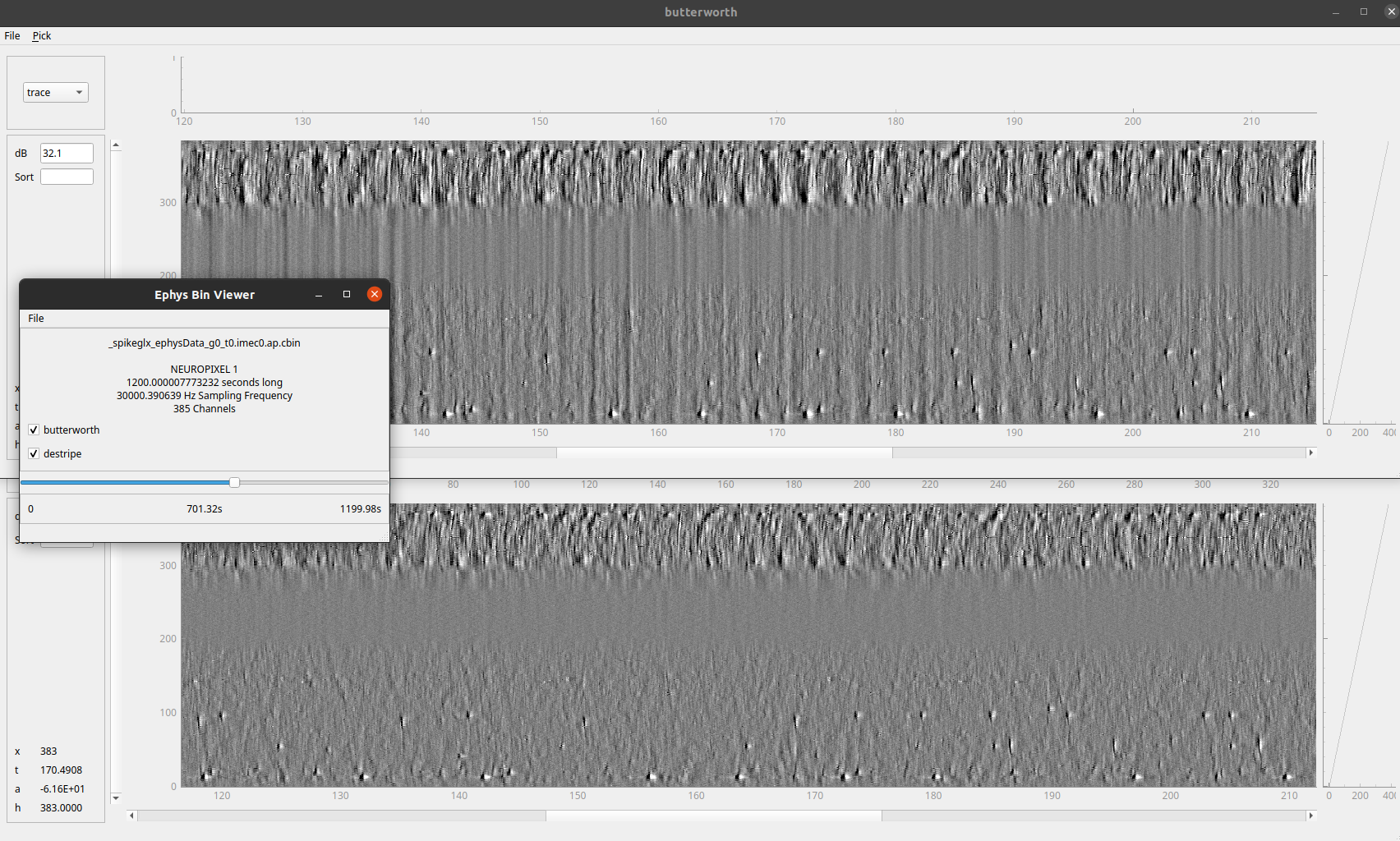
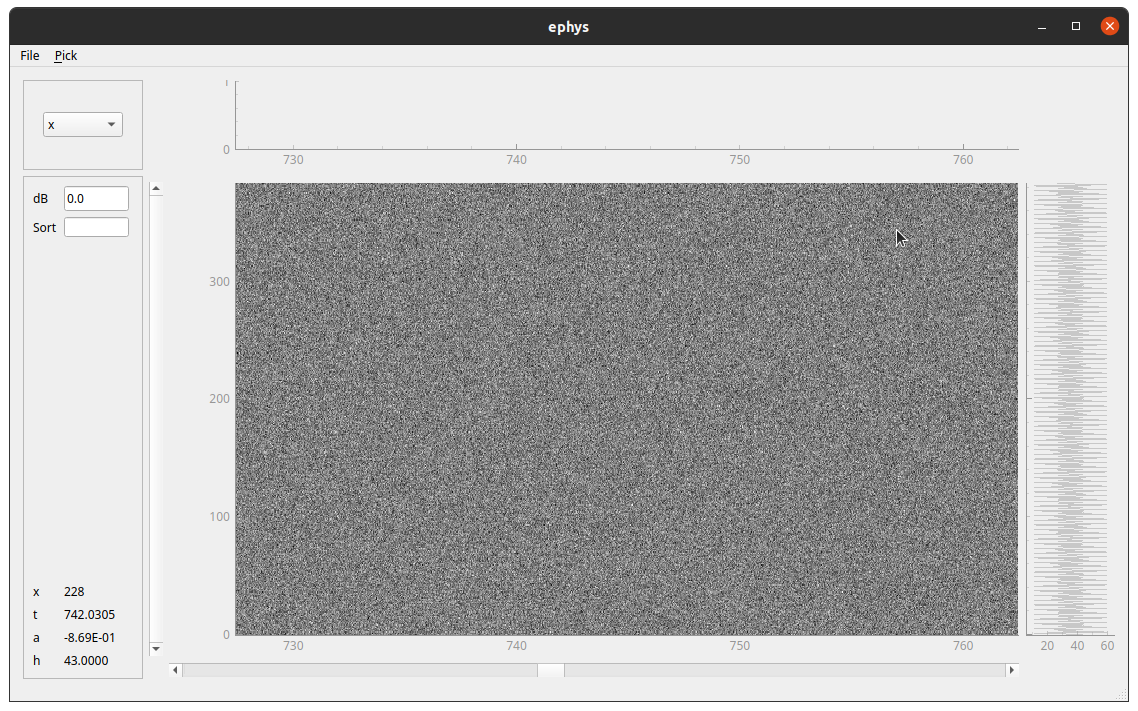
Neuropixel raw data viewer
pip install viewephys
Alternatively, in development mode:
git clone https://github.com/int-brain-lab/viewephys.git
cd viewephys
pip install -e .This is compatible with the IBL environment
Otherwise, you can create a new environment as such:
conda create -n viewephys python=3.12
conda activate viewephysAnd then follow the install instructions above.
ctrl + z: -3dB gainctrl + a: +3dB gainctrl + p: in multi-windows mode, link the displays (pan, zoom and gain)
When the picking mode is enabled (menu pick)
- left button click sets a point
- shift + left button removes a point
- control + left does not wrap on maximum around pick
- space increments the spike group number
Activate your environment and type viewephys, you can then load a neuropixel binary file using the file menu.
Alternatively you can point the viewer to a specific file using the command line:
viewphys -f /path/to/raw.bin# if running ipython, you may have to use the `%gui qt` magic command
import numpy as np
from viewephys.gui import viewephys
nc, ns, fs = (384, 50000, 30000) # this mimics one second of neuropixel data
data = np.random.randn(nc, ns) / 1e6 # volts by default
ve = viewephys(data, fs=fs)Fork and PR.
Pypi Release checklist:
flake8
rm -fR dist
rm -fR build
python setup.py sdist bdist_wheel
twine upload dist/*
#twine upload --repository-url https://test.pypi.org/legacy/ dist/*