This project will no longer be maintained by Intel. Intel has ceased development and contributions including, but not limited to, maintenance, bug fixes, new releases, or updates, to this project. Intel no longer accepts patches to this project.
| Details | |
|---|---|
| Target OS: | Ubuntu* 18.04 LTS |
| Programming Language: | Python* 3.5 |
| Time to Complete: | 30 min |
This reference implementation showcases a health care application by performing pneumonia classification on X-ray images and it results the probability value of disease.
- 6th to 8th generation Intel® Core™ processors with Iris® Pro graphics or Intel® HD Graphics
-
Ubuntu* 18.04 LTS
NOTE: Use kernel versions 4.14+ with this software.
Determine the kernel version with the below uname command.uname -a -
Intel® Distribution of OpenVINO™ toolkit 2020 R3 release
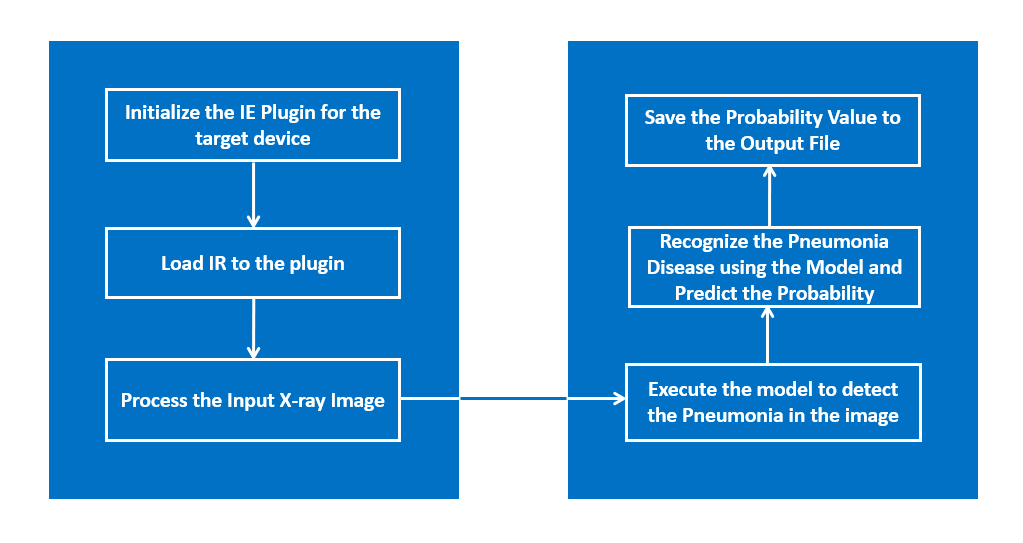
The application uses the Inference Engine included in the Intel® Distribution of OpenVINO™ toolkit. It uses X-ray image as an input source. A trained neural network detects pneumonia probability of preprocessed X-ray image. And it stores the probability and inference time value in a file.
Clone the reference implementation:
sudo apt-get update && sudo apt-get install git
git clone https://github.com/intel-iot-devkit/pneumonia-classification.git
Refer to Install Intel® Distribution of OpenVINO™ toolkit for Linux* to learn how to install and configure the toolkit.
Install the OpenCL™ Runtime Package to run inference on the GPU, as shown in the instructions below. It is not mandatory for CPU inference.
This application uses a pre-trained model, that is provided in the /resources directory. This model is trained using the dataset found in https://data.mendeley.com/datasets/rscbjbr9sj/2, made available under the (CC BY-SA 4.0) license. Instructions on how to train the model can also be found there. This model needs to be passed through the model optimizer to generate the IR (the .xml and .bin files) that will be used by the application.
To install the dependencies of the RI, run the following commands:
cd <path_to_the_pneumonia-classification-python_directory>
./setup.sh
The resources/config.json contains the path to the images that will be used by the application.
The config.json file is of the form name/value pair, image: <path/to/imagefile>
The application can use any number of images for detection.
The application works with any input X-ray input image. For first-use, we recommend using the NORMAL and PNEUMONIA images.
For example:
The config.json would be:
{
"inputs": [
{
"image": "resources/NORMAL/*.jpeg"
},
{
"image": "resources/PNEUMONIA/*.jpeg"
}
]
}
To use any other image, specify the path in config.json file
You must configure the environment to use the Intel® Distribution of OpenVINO™ toolkit one time per session by running the following command:
source /opt/intel/openvino/bin/setupvars.sh
Note: This command needs to be executed only once in the terminal where the application will be executed. If the terminal is closed, the command needs to be executed again.
Change the current directory to the git-cloned application code location on your system:
cd <path_to_the_pneumonia-classification-python_directory>/application
To see a list of the various options:
python3 pneumonia_classification.py -h
To run the application with the needed models:
python3 pneumonia_classification.py -m ../resources/FP32/model.xml
The output images, results.txt and stats.txt files will be saved in the output directory
results.txt: This file contains the probability of pneumonia message and inference time
stats.txt: This file contains the total average inference time
To save the results in a specific directory
python3 pneumonia_classification.py -m ../resources/FP32/model.xml -o <path-to-the-directory_to_save_the_output_files>
A user can specify a target device to run on by using the device command-line argument -d followed by one of the values CPU, GPU,MYRIAD or HDDL
To run with multiple devices use -d MULTI:device1,device2. For example: -d MULTI:CPU,GPU,MYRIAD
Although the application runs on the CPU by default, this can also be explicitly specified through the -d CPU command-line argument:
python3 pneumonia_classification.py -m ../resources/FP32/model.xml -d CPU
-
To run on the integrated Intel® GPU with floating point precision 32 (FP32), use the
-d GPUcommand-line argument:python3 pneumonia_classification.py -m ../resources/FP32/model.xml -d GPUFP32: FP32 is single-precision floating-point arithmetic uses 32 bits to represent numbers. 8 bits for the magnitude and 23 bits for the precision. For more information, click here
-
To run on the integrated Intel® GPU with floating point precision 16 (FP16):
python3 pneumonia_classification.py -m ../resources/FP16/model.xml -d GPUFP16: FP16 is half-precision floating-point arithmetic uses 16 bits. 5 bits for the magnitude and 10 bits for the precision. For more information, click here
To run on the Intel® Neural Compute Stick, use the -d MYRIAD command-line argument:
python3 pneumonia_classification.py -m ../resources/FP16/model.xml -d MYRIAD
Note: The Intel® Neural Compute Stick can only run FP16 models. The model that is passed to the application, through the -m <path_to_model> command-line argument, must be of data type FP16.
To run on the Intel® Movidius™ VPU, use the -d HDDL command-line argument:
python3 pneumonia_classification.py -m ../resources/FP16/model.xml -d HDDL
Note: The Intel® Movidius™ VPU can only run FP16 models. The model that is passed to the application, through the -m <path_to_model> command-line argument, must be of data type FP16.