Thank you for your interest in this project. We regret to inform you that this repository is no longer maintained or actively developed. As a result, we have reached the End of Life (EOL) for this project.
- The project is no longer actively maintained.
- No new features or updates will be added.
- No bug fixes or patches will be provided.
- Issues and pull requests are no longer actively monitored.
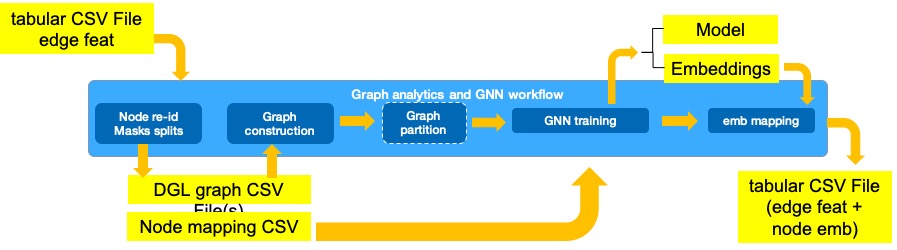
This workflow shows how to run Graph Neural Network (GNN) training on CPUs in single and distributed mode. The workflow reads tabular data, ingests it into graph format, and then uses a GNN to learn embeddings used as rich features in a downstream task.
This workflow is used by the Fraud Detection Reference Kit
Check out more workflow examples and reference implementations in the Dev Catalog.
Graph Neural Networks are effective models for generating node/edge embeddings that can be used as rich features to improve accuracy of downstream tasks. This workflow provides a step-by-step example for how GNNs can be used in fraud detection to extract node embeddings for all entities (credit cards and merchant) based on the graph structure defined by the transactions between them.
General steps:
- Conversion of tabular data into a set of files (nodes.csv, edges.csv and meta.yml). These CSV files form a CSVDataset for ingestion into Deep Graph Library (DGL) graph.
- Training of GNN GraphSAGE model for self-supervised transductive link prediction task. If this is performed on a cluster of machines this training is preceded by graph partitioning.
- Mapping of generated node embeddings into original tabular dataset.
Use cases such as fraud detection, are characterized by class imbalance in their datasets that makes training predictor models directly with those labels difficult. This GNN workflow shows an example of how a self-supervised task can be formulated (instead of using the hard imbalanced labels) to learn entity features that capture the graph structure for use by a downstream predictor model such as XGBoost. In this workflow, the self-supervised task consists of link prediction where the edges in the graph are used as positive examples and non-existent edges as negative examples.
GNN workflow ingests tabular data where each row corresponds to a transaction between two types of entities: cards and merchants, and generates a graph where entities are the nodes (that are featureless), and transactions constitute the edges with the associated transaction feature attributes.
The GNN model consists of a learnable embedding layer followed by an encoder, implemented as a 2-layer GraphSAGE model, and a decoder, implemented as a 3-layer multilayer perceptron (MLP) with a single output for link prediction. During training, positive and negative neighbor sampling is used to generate the training examples. We use a Receiver Operating Characteristic Area Under Curve score (ROC AUC) as the metric to evaluate the quality of the embeddings in predicting if two entities should be connected. The ultimate measure of how useful these embeddings are in predicting fraud needs to be measured by the downstream predictor model, since this workflow is not using the fraud labels directly.
After several epochs of training, we run GNN inference on the entire graph without neighbor sampling and use the last layer activations generated by the model as node embeddings for nodes of the graph. These embeddings can be mapped to the entities in the tabular data input and used as node features for a downstream prediction task.
There are workflow-specific hardware and software setup requirements depending on how the workflow is run. Bare metal development system and Docker image running locally have the same system requirements.
| Recommended Hardware | Precision |
|---|---|
| Intel® 1st, 2nd, 3rd, and 4th Gen Xeon® Scalable Performance processors | FP32 |
For distributed training a high-speed fabric across nodes (e.g., OPA, Mellanox) is recommended.
Workflow has been tested on OS Rocky Linux v8.7 and Ubuntu 20.04
Create a working directory for the workflow and clone the graph-neural-networks-and-analytics repository into your working directory.
mkdir ~/work && cd ~/work
git clone https://github.com/intel/graph-neural-networks-and-analytics
cd graph-neural-networks-and-analytics
git checkout tags/v1.0-beta
export WORKSPACE=$PWD
The input to this workflow is tabular data in CSV format where each row corresponds to a transaction between two entities. In the case of the IBM/tabformer dataset used by Fraud Detection Reference Kit it consists of credit card transaction. Each transaction includes the IDs of the entities involved (Card and Merchant) and the edge features (amount of the transaction, date, etc.).
There are two options for processing the input data:
If you are running this workflow as part of the Fraud Detection Reference Kit the input to this workflow will be the output of the data preprocesssing (featurized edge data) stage.
OR
If you want to run this workflow in standalone mode, download the synthetic credit card transaction dataset IBM/tabformer and use the utility script below to featurize the data using pandas.
#run this script to create conda environment
./script/build_dgl1_env.sh
conda activate dgl1.0
#path to downloaded data. Include filename
DATA_IN=< YOUR PATH TO >/card_transaction.v1.csv
#path for processed_data.csv. Include filename
PROCESSED_DATA=< PATH TO SAVE >/processed_data.csv
#read raw data, perform edge featurization and generate CSVDataset files for ingesting graph
./script/run_data_prep.sh $DATA_IN $PROCESSED_DATAYou can bring your own tabular transaction dataset to be used with this workflow. For the workflow to be able to model the graph correctly user needs to update the
tabular2graph.yaml to include the column names for the node IDs, the edge types (triplets of (src_types, edge_type, dst_type), the column name for the label, list of column names for edge features, etc.
Please refer to the workflow-config.yaml for a detailed description of input configurations.
These are the minimum requiremenets on tabular dataset:
- CSV file including header with column names. User will use the
tabular2graph.yamlto indicate what are the column names corresponding to entity IDs, label, features... - Each row should represent a link in the graph thus including at minimum the IDs for the two connecting entities and a column indicating the train/val/test split using 0,1,2 values respectively.
- The number of node types and edge types is not constrained and will be provided by user in the "tabular2graph.yaml"
- label, column (edge) features are optional and will be included in the CSVDataset if provided in the
tabular2graph.yaml - Because current workflow is useful to model unsupervised link prediction tasks there is currently no support for other type of graph inputs such as multi graph.
This workflow supports running in different ways:
- Run single node bare metal
- Run Single node Using Docker
- Run Bare metal on a cluster of machines
Please ensure you meet the below prerequisites based on your selection
- Our examples use the
condapackage and environment on your local computer. If you don't already havecondainstalled, see the Conda Linux installation instructions. - Have numactl installed on your system. You can use
sudo apt-get install numactl(Ubuntu)dnf install numactl(CentOS)
You'll need to install Docker Engine on your development system. Note that while Docker Engine is free to use, Docker Desktop may require you to purchase a license. See the Docker Engine Server installation instructions for details.
-
Our examples use the
condapackage and environment on your local computer. If you don't already havecondainstalled, see the Conda Linux installation instructions. -
Before you can run in a distributed cluster, you need to configure passwordless ssh access across machines and have a distributed file system so the conda environment, data and files can be accessed across multiple machines. See these linked documents on how to set up passwordless ssh and set up Distributed File System.
-
Have numactl installed on your system. You can use
sudo apt-get install numactl(Ubuntu)dnf install numactl(CentOS)
This GNN workflow can be configured by the user using yaml configuration files and it supports running in different ways:
- Run single node bare metal
- Run Single node Using Docker
- Run Bare metal on a cluster of machines
The selection between these different modes can be done in the workflow-config.yaml.
In these sections you will find instructions on how to update the configuration yaml files to run this workflow.
workflow-config.yaml is the main configuration file for the user to specify:
- Runtime environment (i,e number of nodes in cluster, IPs, bare metal/docker, ...)
- Directories for inputs, outputs and configuration files
- Configure what stages of the workflow to execute. A user may run all stages the first time but may want to skip building or partitioning a graph in later training experiments to save time.
Please refer to the workflow-config.yaml for a detailed description of input configurations.
In model-training.yaml user can specify:
- Dataloader, sampler and model parameters (i,e batch size, sampling fanout, learning rate)
- Training hyperparamets (i,e number of epochs)
- DGL specific parameters for distributed training
Please refer to the model-training.yaml for a detailed description of input configurations.
Once the prerequisits have been met and the 3 configuration files have been updated to execute the workflow:
./run-workflow.sh ./configs/workflow-config.yamlThe successful execution of this stage will create the below contents under ${env_tmp_path} directory specified in workflow-config.yaml:
./sym_tabformer_hetero_CSVDataset/
├── edges_0.csv
├── edges_1.csv
├── meta.yaml
├── nodes_0.csv
└── nodes_1.csv
The successful execution of this stage will create the below contents under ${env_tmp_path}/partitions directory:
tabformer_2parts/
├── emap.pkl
├── nmap.pkl
├── part0
│ ├── edge_feat.dgl
│ ├── graph.dgl
│ └── node_feat.dgl
├── part1
│ ├── edge_feat.dgl
│ ├── graph.dgl
│ └── node_feat.dgl
└── tabformer_full_homo.json
The successful training will show the epoch times as it progresses along with roc_auc scores. The training logs are saved under:
#the single node logs can be found
${WORKSPACE}/logs
#the distributed training log can be found at:
${WORKSPACE}/logs_distThe "map_save" stage generates a CSV file combining the input tabular data and the node embeddings generated by this GNN workflow. In the case of Tabformer datast this file will have 154 features per transaction and can be used for the fraud prediction downstream task. For an example of fraud detection with XGBoost using these gnn_features, refer to Fraud Detection Reference Kit.
The mappings happening in this stage are:
- the mapping from the local to global graph IDs (node/edge ID's) and
- the mapping between global graph ID's and the card and merchant ID's in the tabular dataset.
The successful mapping of the embeddings will show the following details
Loading embeddings from file and adding to preprocessed CSV file
CSV output shape: (24198836, 154)
Time to append node embeddings to edge features CSV <this will vary based on system>
The output file (~35GB) can be found on your host system's output directory indicated by ${env_out_path} in workflow-config.yaml:
#resulting file with edge features and node embeddings
$OUT_DIR/tabular_with_gnn_emb.csv
We have seen how tabular data such as credit card transactions can be modeled as a graph and how GNNs can be used to generate node embeddings used to boost accuracy of downstream tasks. In addition we have shown how to bring your own tabular data and perform this GNN training on a container or on bare metal as a single node or multiple nodes by modifying a set of configuration files.
For more information about Graph Neural Networks and Analytics workflow or to read about other relevant workflow examples, see these guides and software resources:
- Distributed training uses Pytorch with GLOO backend. If it gives a Connection Refused RuntimeError from third_party/gloo you may try adding
--extra_envs GLOO_SOCKET_IFNAME=eth0in run_dist_train.sh after launch.py. Replaceeth0above with the right network interface in your system. - If distributed training gets interrupted and cannot restart training successfully afterwards you may need to manually kill python processes in the remote machines.
- If you see a TargetEncoder python import error when running run_data_prep.sh you can install it into your dgl1.0 conda environment using
conda install -c conda-forge category_encoders. - If you get a Permission denied error when running a bash script such as
./script/run_build_graph.sh, it can be fixed bychmod +x ./script/run_build_graph.sh
*Other names and brands may be claimed as the property of others. Trademarks.