This is an official implementation of “Adapting Visual-Language Models for Generalizable Anomaly Detection in Medical Images” with PyTorch, accepted by CVPR 2024 (Highlight).
If our work is helpful for your research, please consider citing:
@inproceedings{huang2024adapting,
title={Adapting Visual-Language Models for Generalizable Anomaly Detection in Medical Images}
author={Huang, Chaoqin and Jiang, Aofan and Feng, Jinghao and Zhang, Ya and Wang, Xinchao and Wang, Yanfeng},
booktitle={IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)},
year={2024}
}
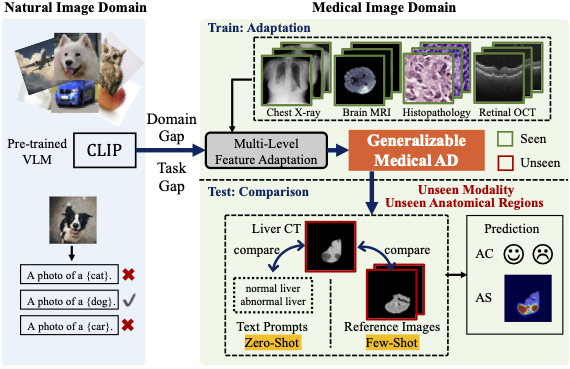
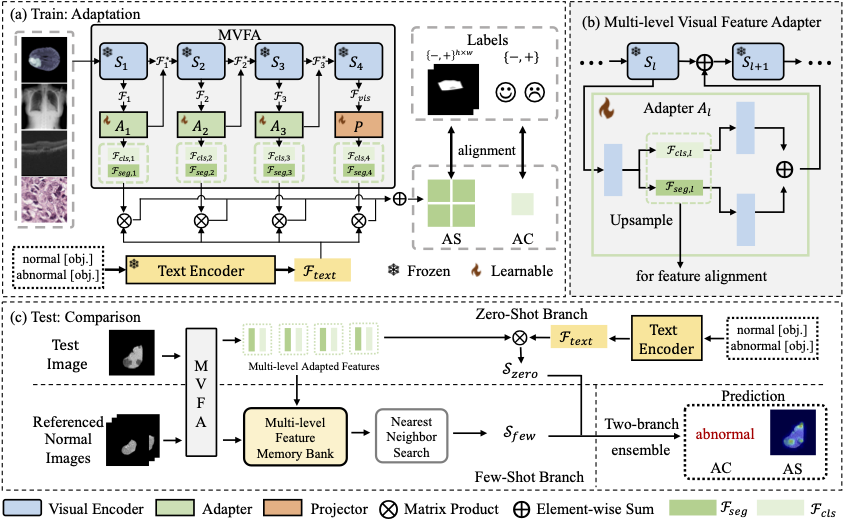
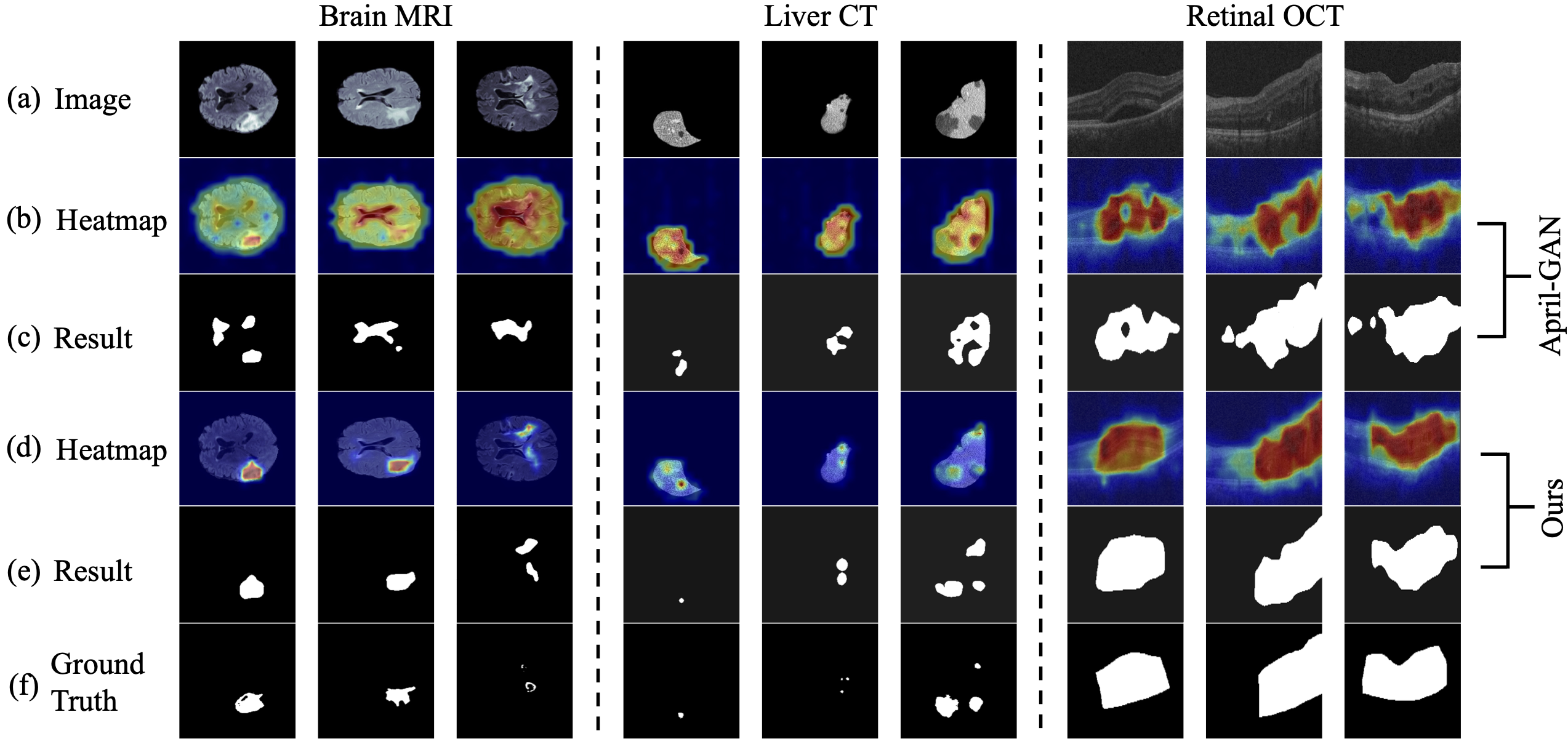
Abstract: Recent advancements in large-scale visual-language pre-trained models have led to significant progress in zero-/few-shot anomaly detection within natural image domains. However, the substantial domain divergence between natural and medical images limits the effectiveness of these methodologies in medical anomaly detection. This paper introduces a novel lightweight multi-level adaptation and comparison framework to repurpose the CLIP model for medical anomaly detection. Our approach integrates multiple residual adapters into the pre-trained visual encoder, enabling a stepwise enhancement of visual features across different levels. This multi-level adaptation is guided by multi-level, pixel-wise visual-language feature alignment loss functions, which recalibrate the model’s focus from object semantics in natural imagery to anomaly identification in medical images. The adapted features exhibit improved generalization across various medical data types, even in zero-shot scenarios where the model encounters unseen medical modalities and anatomical regions during training. Our experiments on medical anomaly detection benchmarks demonstrate that our method significantly surpasses current state-of-the-art models, with an average AUC improvement of 6.24% and 7.33% for anomaly classification, 2.03% and 2.37% for anomaly segmentation, under the zero-shot and few-shot settings, respectively.
Keywords: Anomaly Detection, Medical Images
- python >= 3.8.5
- pytorch >= 1.10.0
- torchvision >= 0.11.1
- numpy >= 1.19.2
- scipy >= 1.5.2
- kornia >= 0.6.1
- pandas >= 1.1.3
- opencv-python >= 4.5.4
- pillow
- tqdm
- ftfy
- regex
Single NVIDIA GTX 3090
-
Download and put it under
CLIP/ckptfolder -
MVFA:
- few-shot: https://drive.google.com/file/d/1bV1yzPxJarTRfd8liMIwyHcGywTTEL2k/view?usp=sharing
- zero-shot: https://drive.google.com/file/d/1nGhcK32CrkgTR5Rav6rNfptUHaASfRnU/view?usp=sharing
Download and put it under
ckptfolder. Please also unzip the ckpt files:unzip few-shot.zip unzip zero-shot.zip
-
(optional) Follow the BMAD to apply for permission to download the relevant dataset. After extracting the data, reorganize the data benchmark according to the guidelines provided in our Appendix A.
-
We also provide the pre-processed benchmark. Please download the following dataset
- Liver: https://drive.google.com/file/d/1xriF0uiwrgoPh01N6GlzE5zPi_OIJG1I/view?usp=sharing
- Brain: https://drive.google.com/file/d/1YxcjcQqsPdkDO0rqIVHR5IJbqS9EIyoK/view?usp=sharing
- HIS: https://drive.google.com/file/d/1hueVJZCFIZFHBLHFlv1OhqF8SFjUVHk6/view?usp=sharing
- RESC: https://drive.google.com/file/d/1BqDbK-7OP5fUha5zvS2XIQl-_t8jhTpX/view?usp=sharing
- OCT17: https://drive.google.com/file/d/1GqT0V3_3ivXPAuTn4WbMM6B9i0JQcSnM/view?usp=sharing
- ChestXray: https://drive.google.com/file/d/15DhnBAk-h6TGLTUbNLxP8jCCDtwOHAjb/view?usp=sharing
-
Place it within the master directory
dataand unzip the dataset.tar -xvf Liver.tar.gz tar -xvf Brain.tar.gz tar -xvf Histopathology_AD.tar.gz tar -xvf Retina_RESC.tar.gz tar -xvf Retina_OCT2017.tar.gz tar -xvf Chest.tar.gz
After the preparation work, the whole project should have the following structure:
code
├─ ckpt
│ ├─ few-shot
│ └─ zero-shot
├─ CLIP
│ ├─ bpe_simple_vocab_16e6.txt.gz
│ ├─ ckpt
│ │ └─ ViT-L-14-336px.pt
│ ├─ clip.py
│ ├─ model.py
│ ├─ models.py
│ ├─ model_configs
│ │ └─ ViT-L-14-336.json
│ ├─ modified_resnet.py
│ ├─ openai.py
│ ├─ tokenizer.py
│ └─ transformer.py
├─ data
│ ├─ Brain_AD
│ │ ├─ valid
│ │ └─ test
│ ├─ ...
│ └─ Retina_RESC_AD
│ ├─ valid
│ └─ test
├─ dataset
│ ├─ fewshot_seed
│ │ ├─ Brain
│ │ ├─ ...
│ │ └─ Retina_RESC
│ ├─ medical_few.py
│ └─ medical_zero.py
├─ loss.py
├─ prompt.py
├─ readme.md
├─ train_few.py
├─ train_zero.py
└─ utils.py
python test_few.py --obj $target-object --shot $few-shot-number
For example, to test on the Brain MRI with k=4, simply run:
python test_few.py --obj Brain --shot 4
python train_few.py --obj $target-object --shot $few-shot-number
For example, to train on the Brain MRI with k=4, simply run:
python train_few.py --obj Brain --shot 4
Results of zero-shot anomaly detection and localization:
| AUC (%) | Detection | Localization | ||
| Zero-shot | Paper | Inplementation | Paper | Inplementation |
| HIS | 77.90 | 76.90 | - | - |
| ChestXray | 71.11 | 71.11 | - | - |
| OCT17 | 95.40 | 95.40 | - | - |
| BrainMRI | 78.63 | 79.80 | 90.27 | 89.68 |
| LiverCT | 76.24 | 81.18 | 97.85 | 97.93 |
| RESC | 83.31 | 88.99 | 92.05 | 90.44 |
| Average | 80.43 | 82.23 | 93.39 | 92.68 |
Results of few-shot anomaly detection and localization with k=4:
| AUC (%) | Detection | Localization | ||
| 4-shot | Paper | Inplementation | Paper | Inplementation |
| HIS | 82.71 | 82.71 | - | - |
| ChestXray | 81.95 | 81.95 | - | - |
| OCT17 | 99.38 | 99.38 | - | - |
| BrainMRI | 92.44 | 92.31 | 97.30 | 97.30 |
| LiverCT | 81.18 | 81.18 | 99.73 | 99.69 |
| RESC | 96.18 | 96.18 | 98.97 | 98.97 |
| Average | 88.97 | 88.95 | 98.67 | 98.65 |
We borrow some codes from OpenCLIP, and April-GAN.
If you have any problem with this code, please feel free to contact huangchaoqin@sjtu.edu.cn and stillunnamed@sjtu.edu.cn.