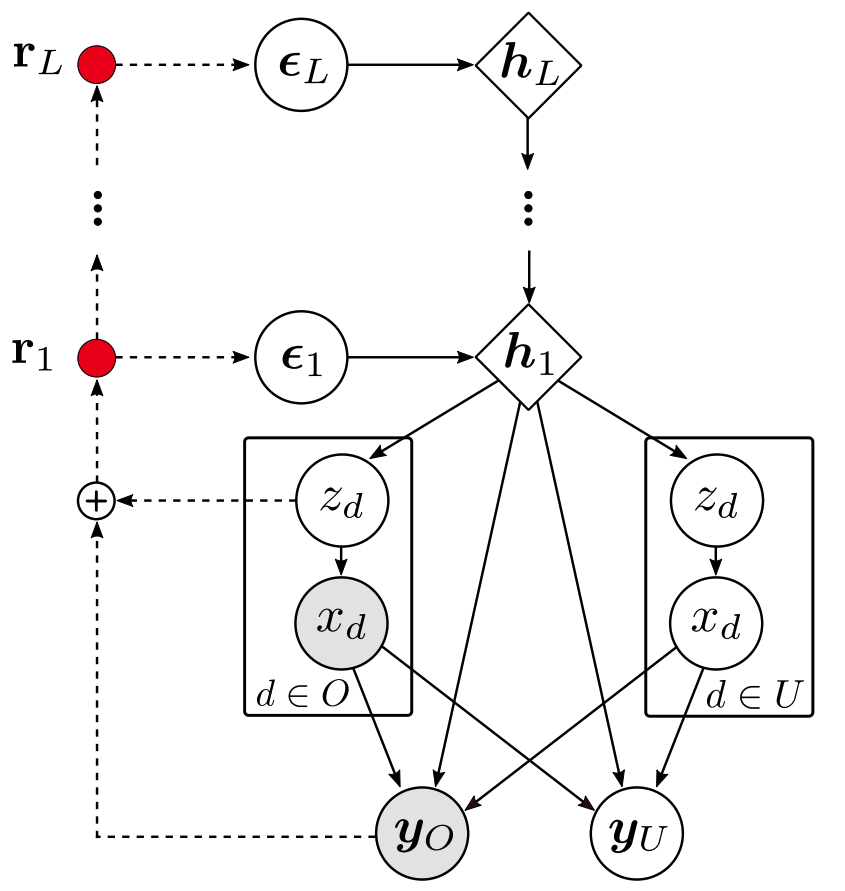
This repository contains the official Pytorch implementation of the Hierarchical Hamiltonian VAE for Mixed-type Data (HH-VAEM) model and the sampling-based feature acquisition technique presented in the paper Missing Data Imputation and Acquisition with Deep Hierarchical Models and Hamiltonian Monte Carlo. HH-VAEM is a Hierarchical VAE model for mixed-type incomplete data that uses Hamiltonian Monte Carlo with automatic hyper-parameter tuning for improved approximate inference. The repository contains the implementation and the experiments provided in the paper.
Please, if you use this code, cite the preprint using:
@inproceedings{peis2022missing,
abbr={NeurIPS},
title={Missing Data Imputation and Acquisition with Deep Hierarchical Models and Hamiltonian Monte Carlo},
author={Peis, Ignacio and Ma, Chao and Hern{\'a}ndez-Lobato, Jos{\'e} Miguel},
booktitle={Advances in Neural Information Processing Systems},
volume={35},
year={2022},
}
The installation is straightforward using the following instruction, that creates a conda virtual environment named HH-VAEM using the provided file environment.yml:
conda env create -f environment.yml
The project is developed in the recent research framework PyTorch Lightning. The HH-VAEM model is implemented as a LightningModule that is trained by means of a Trainer. A model can be trained by using:
# Example for training HH-VAEM on Boston dataset
python train.py --model HHVAEM --dataset boston --split 0
This will automatically download the boston dataset, split in 10 train/test splits and train HH-VAEM on the training split 0. Two folders will be created: data/ for storing the datasets and logs/ for model checkpoints and TensorBoard logs. The variable LOGDIR can be modified in src/configs.py to change the directory where these folders will be created (this might be useful for avoiding overloads in network file systems).
The following datasets are available:
- A total of 10 UCI datasets:
avocado,boston,energy,wine,diabetes,concrete,naval,yatch,bankorinsurance. - The MNIST datasets:
mnistorfashion_mnist. - More datasets can be easily added to
src/datasets.py.
For each dataset, the corresponding parameter configuration must be added to src/configs.py.
The following models are also available (implemented in src/models/):
HHVAEM: the proposed model in the paper.VAEM: the VAEM strategy presented in (Ma et al., 2020) with Gaussian encoder (without including the Partial VAE).HVAEM: A Hierarchical VAEM with two layers of latent variables and a Gaussian encoder.HMCVAEM: A VAEM that includes a tuned HMC sampler for the true posterior.- For MNIST datasets (non heterogeneous data), use
HHVAE,VAE,HVAEandHMCVAE.
By default, the test stage will be executed at the end of the training stage. This can be cancelled with --test 0 for manually running the test using:
# Example for testing HH-VAEM on Boston dataset
python test.py --model HHVAEM --dataset boston --split 0
which will load the trained model to be tested on the boston test split number 0. Once all the splits are tested, the average results can be obtained using the script in the run/ folder:
# Example for obtaining the average test results with HH-VAEM on Boston dataset
python test_splits.py --model HHVAEM --dataset boston
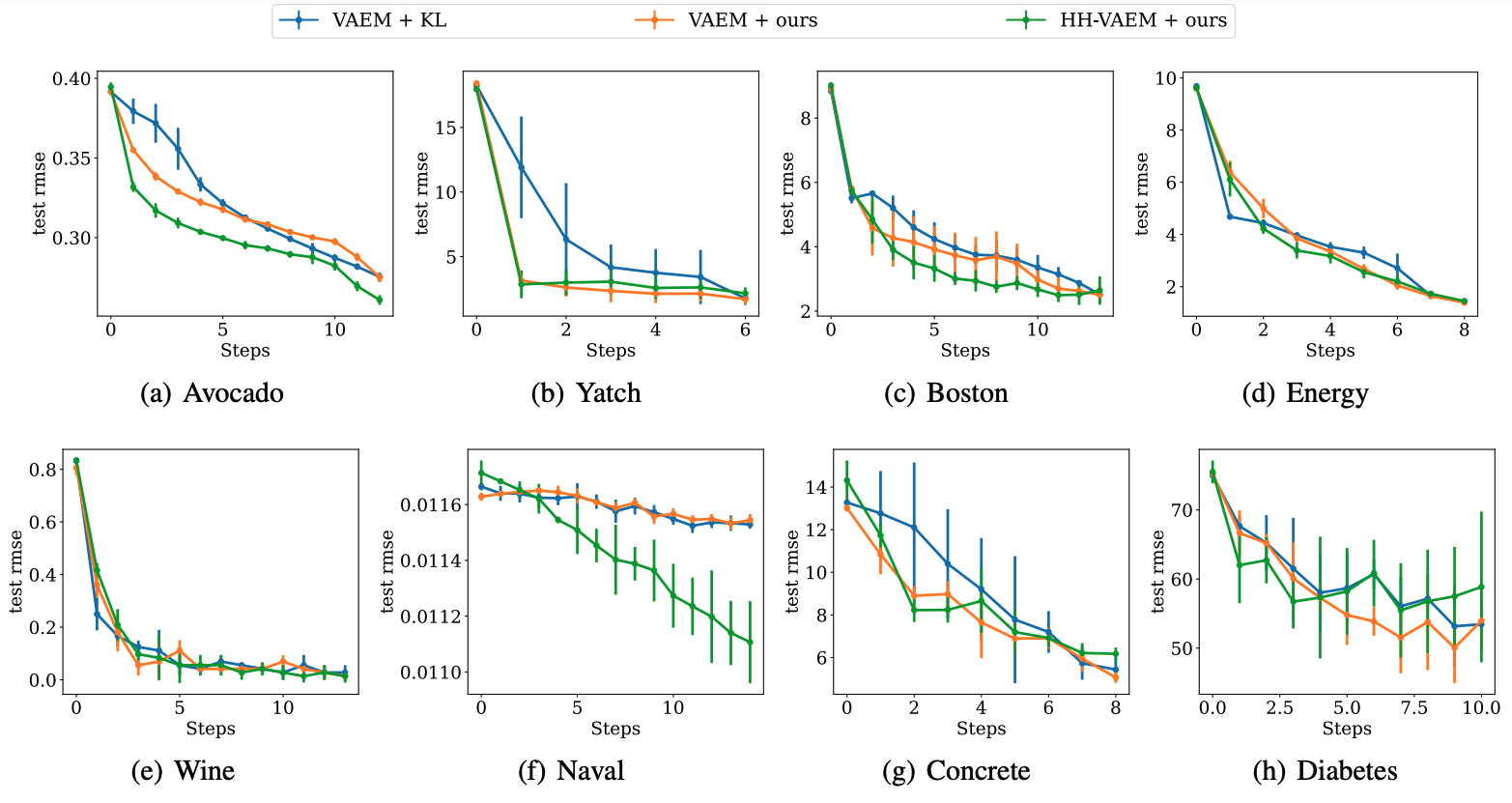
The SAIA experiment in the paper can be executed using:
# Example for running the SAIA experiment with HH-VAEM on Boston dataset
python active_learning.py --model HHVAEM --dataset boston --method mi --split 0
Once this is executed on all the splits, you can plot the SAIA error curves using the scripts in the run/ folder:
# Example for running the SAIA experiment with HH-VAEM on Boston dataset
python active_learning_plots.py --models VAEM HHVAEM --dataset boston
You can also try running the inpainting experiment using:
# Example for running the inpainting experiment using CelebA:
python inpainting.py --models VAE HVAE HMCVAE HHVAE --dataset celeba --split 0
which will stack a row of inpainted images for each of the given models, after two rows with the original and observed images, respectively.
Use the --help option for documentation on the usage of any of the mentioned scripts.
For further information: ipeis@tsc.uc3m.es