This repository contains a Python package for running HMC with Pytorch, including automatic optimization of its hyperparameters. You will be able to i) sample from any distribution, given as a unnormalized target, and ii) automatically tune the HMC hyperparameters to improve the efficiency in exploring the density.
For further details about the algorithm, see Section 3.5 of our paper, where we adapted the HMC tuning for improving the inference in a Hierarchical VAE for mixed-type partial data. Original idea for optimizing HMC via Variational Inference can be found here. If you refer to this algorithm, please consider citing both works. If you use this code, please cite:
@article{peis2022missing,
title={Missing Data Imputation and Acquisition with Deep Hierarchical Models and Hamiltonian Monte Carlo},
author={Peis, Ignacio and Ma, Chao and Hern{\'a}ndez-Lobato, Jos{\'e} Miguel},
journal={arXiv preprint arXiv:2202.04599},
year={2022}
}
The installation is straightforward using the following instruction, that creates a conda virtual environment named HMCTuning using the provided file environment.yml:
conda env create -f environment.yml
For an extended usage guide, check notebooks/usage.ipynb. For a basic usage, continue reading here. An HMC object can be created as in the following example:
from examples.distributions import *
from examples.utils import *
# Load the log probability function of MoG, and the initial proposal
logp = get_logp('gaussian_mixture')
mu0, var0 = initial_proposal('gaussian_mixture') # [0, 0], [0.01, 0.01]
# Create the HMC object
hmc = HMC(dim=2, logp=logp, T=5, L=5, chains=1000, chains_sksd=30, mu0=mu0, var0=var0, vector_scale=True)
where:
-
dimis anintwith the dimension of the target space. -
logpis aCallable(function) that returns the log probability$\log p(\mathbf{x})$ for an input$\mathbf{x}$ . -
Tis anintwith the length of the chains. -
Lis anintwith the number of Leapfrog steps. -
chainsis anintwith the number of parallel chains used for each optimization step. -
chains_sksdis anintwith the number of parallel chains used independently for computing the SKSD discrepancy within each optimization step. -
mu0is a(bath_size, D)tensor with the means of the Gaussian initial proposal. -
var0is a(bath_size, D)tensor with the variances of the Gaussian initial proposal.
For sampling from the created HMC object, just call:
samples, chains = hmc.sample(N)
Your final N samples will be stored in samples, and, if needed, you can inspect the full chains.
To train the HMC hyperparameters, call:
hmc.fit(steps=100)
This will run the gradient-based optimization algorithm that tunes the hyperparameters using Variational Inference.
In the following gifs you can observe two simple examples on how effective is the training algorithm for wave-shaped (left) and dual-mooon (right) densities. Horizontal scaling is automatically increased during training to inflate the proposal for covering the density.
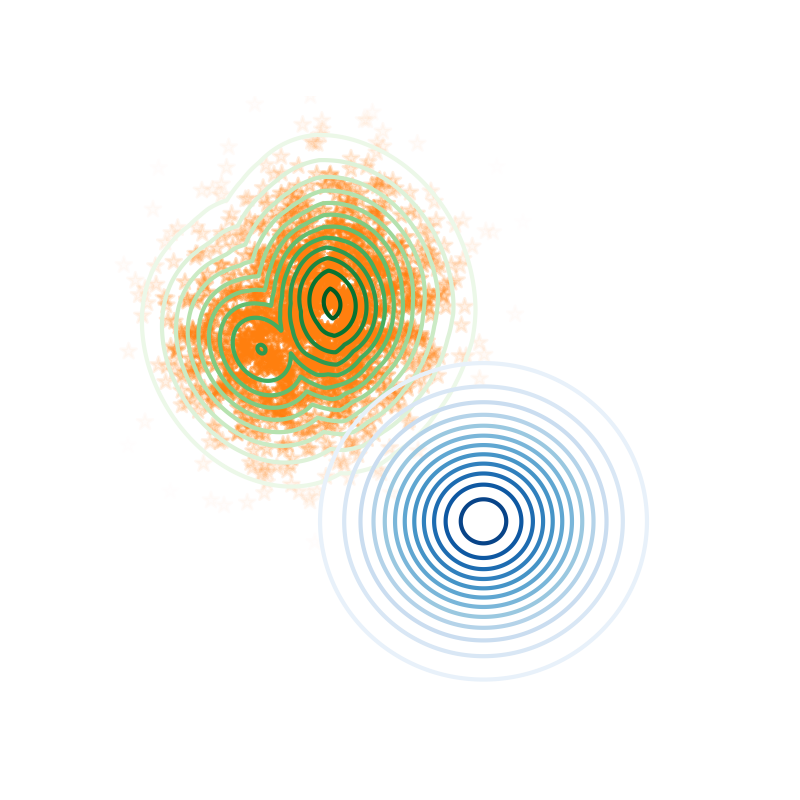
We used our method for improving the inference of advanced Variational Autoencoders like the one presented in our paper. In the following Figure, an illustrative example for a simple VAE is included: when training the VAE parameters jointly with the HMC hyperparameters, the multimodal true posterior (green) is successfully explored with HMC samples (orange) using the Gaussian proposal provided by the encoder (blue).
Use the --help option for documentation on the usage of any of the mentioned scripts.
For further information: ipeis@tsc.uc3m.es