Neural-Fingerprints-Pytorch
Description
Pytorch implementation of Convolutional Networks on Graphs for Learning Molecular Fingerprints. Generate data-driven Molecular Fingerprints from SMILES.
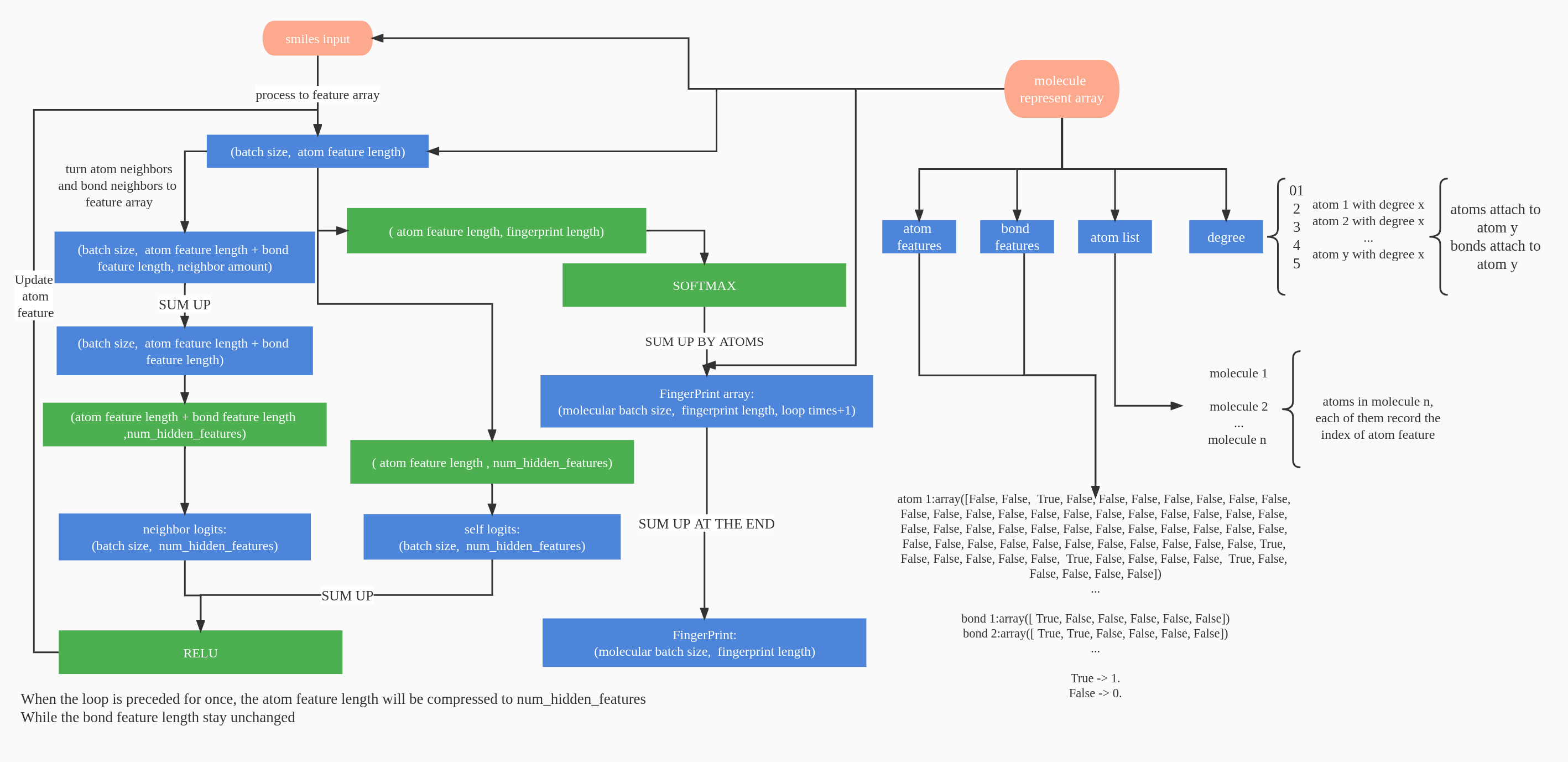 The available structure of another kind of Molecular Fingerprints Networks in convnet_v2.py is shown as follow:
The available structure of another kind of Molecular Fingerprints Networks in convnet_v2.py is shown as follow:
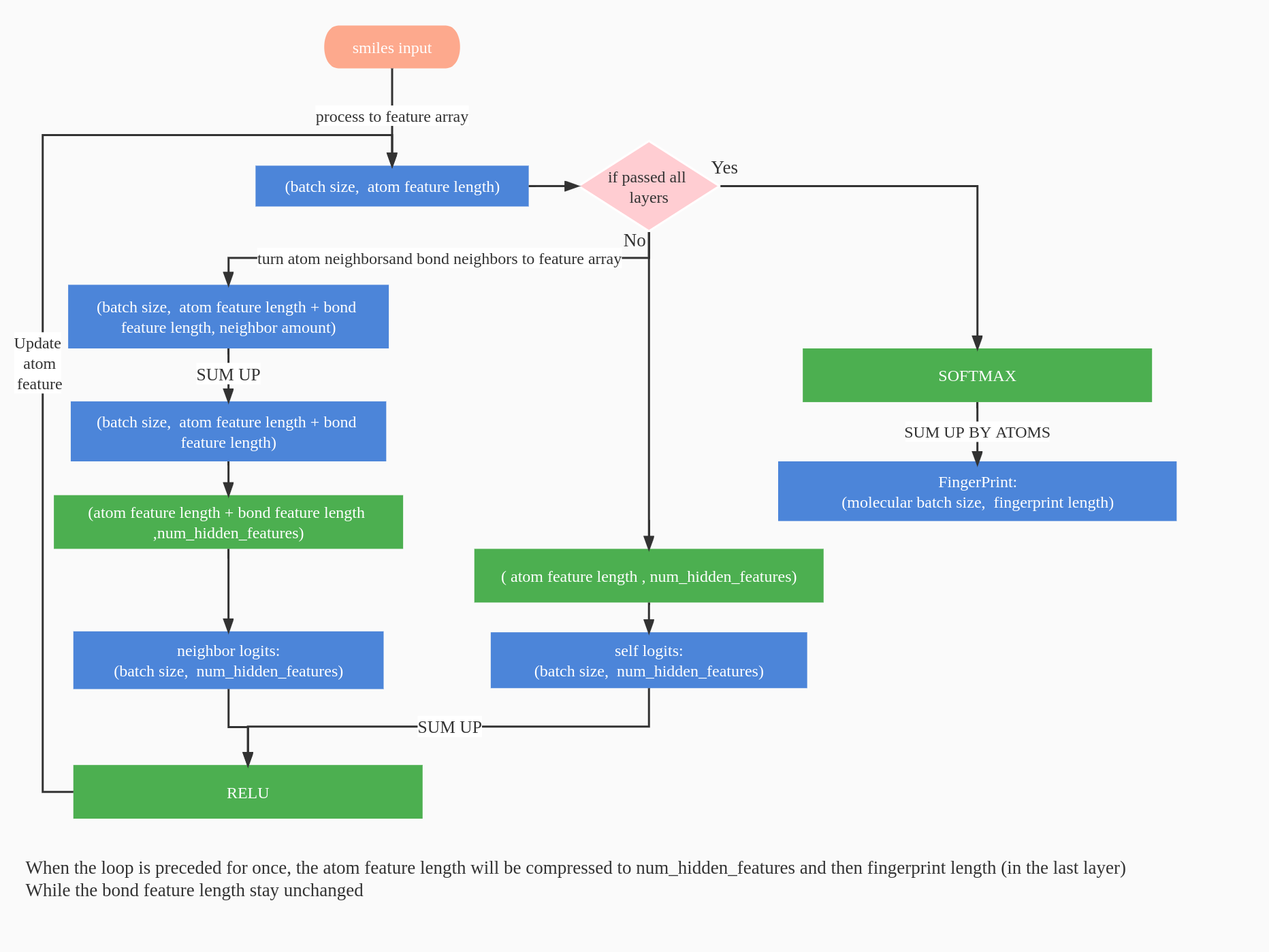
Usage
regression.py is an example script to do regression work using neural fingerprints.
- there are a convolutional network for fingerprints generation and a basic ANN network in these scripts, they can be generated by
from neural_fingerprints.convnet import NeuralConvNetwork
from neural_fingerprints.deepnet import DeepNetwork
model_fp = NeuralConvNetwork(**conv_params)
model_deep = DeepNetwork(**ann_params)- the params for network incloud these contents, which is familiar with those by HIPS:
conv_params = {
fp_length=50, # output of conv network
fp_depth=4, # depth of conv network
conv_width=20, # node numbers of hidden layer
h1_size=100
}
ann_params = {
layer_sizes=[model_params['fp_length'],
model_params['h1_size']],
normalize=True
}Other implementations
- by HIPS using autograd, the direct reference for writing these script
- by debbiemarkslab using theano
- by keiserlab using keras
- by DeepChem using tensorflow
Dependencies
- RDKit
- Pytorch = 1.x
- NumPy