This repository contains a user-friendly PyTorch implementation of StarDist 3D for nuclei segmentation based on:
-
Martin Weigert, Uwe Schmidt, Robert Haase, Ko Sugawara, and Gene Myers.
Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy. The IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, Colorado, March 2020 -
and Stardist PyTorch implementation by [hthierno] https://github.com/hthierno/pytorch-stardist
| Input images | StarDist3D predictions |
|---|---|
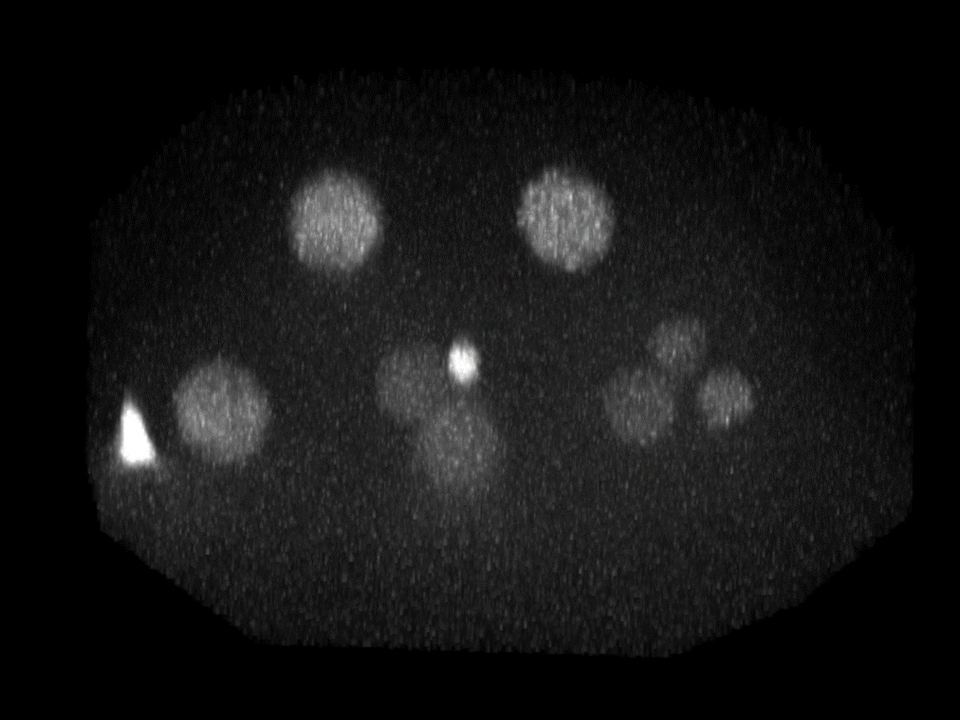 |
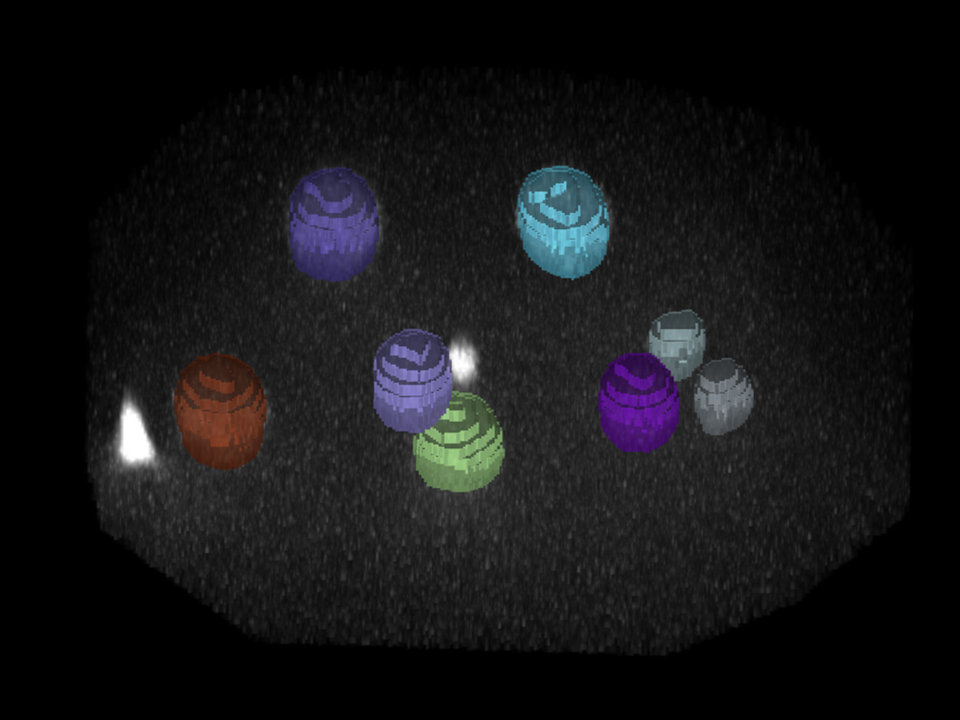 |
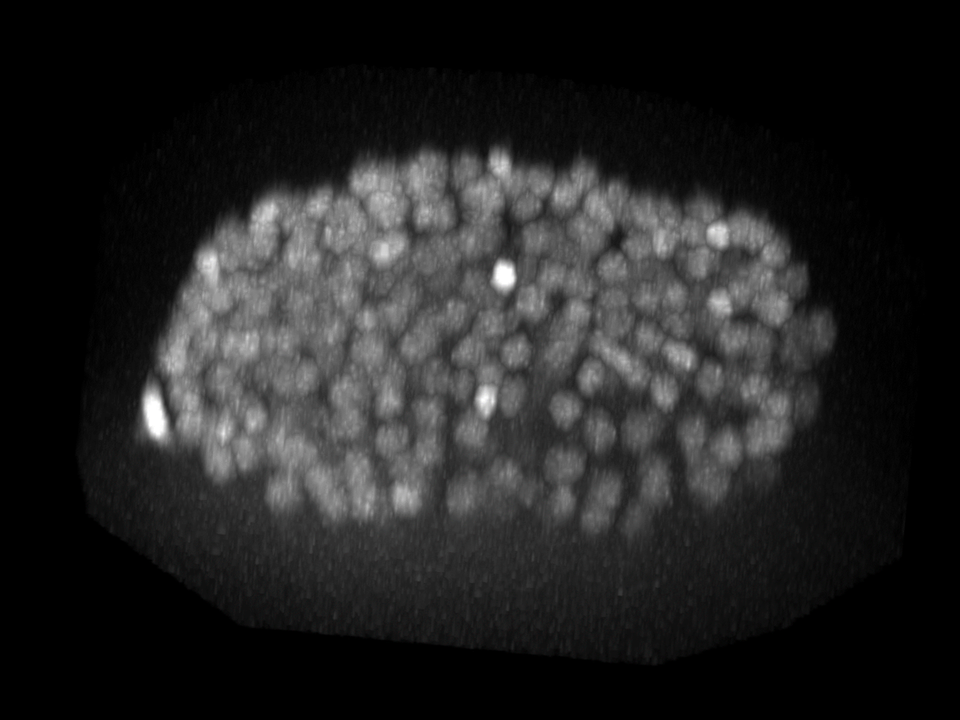 |
 |
You should have a C++ compiler installed as this code relies on C/C++ extensions that need to be compiled. You may follow these steps to install Stardist 3D:
-
Download the repo:
git clone https://github.com/itarampoulous/StarDist3D -
Create a python 3.7.9 conda environment with cuda toolkit for GPU-based computing:
conda create --name "environment_name" python=3.7.9 cudatoolkit -
Activate the conda environment:
conda activate "environment_name" -
Navigate to the directory (StarDist3D) containing the requirements.txt file and install the dependencies:
pip install -r ./requirements.txt -
Install the package by running from the same directory containing the setup.py script:
pip install .
Training using a training configuration file (see example):
trainStardist3D --config path/to/training_configuration.yml
Note that the dataset has to have the following structure:
dataset
── train
├── images
├── masks
── val (optional, if you want to evaluate during training)
├── images
├── masks
Prediction using a prediction configuration file (see example):
predictStardist3D --config path/to/prediction_configuration.yml
Evaluation using an evaluation configuration file (see example):
evaluateStardist3D --config path/to/evaluation_configuration.yml
Examples of evaluation metrics generated by evaluateStardist3D on C. elegans embryo:
| Evaluation on ground truth nuclei centroids | Evaluation on ground truth masks |
|---|---|
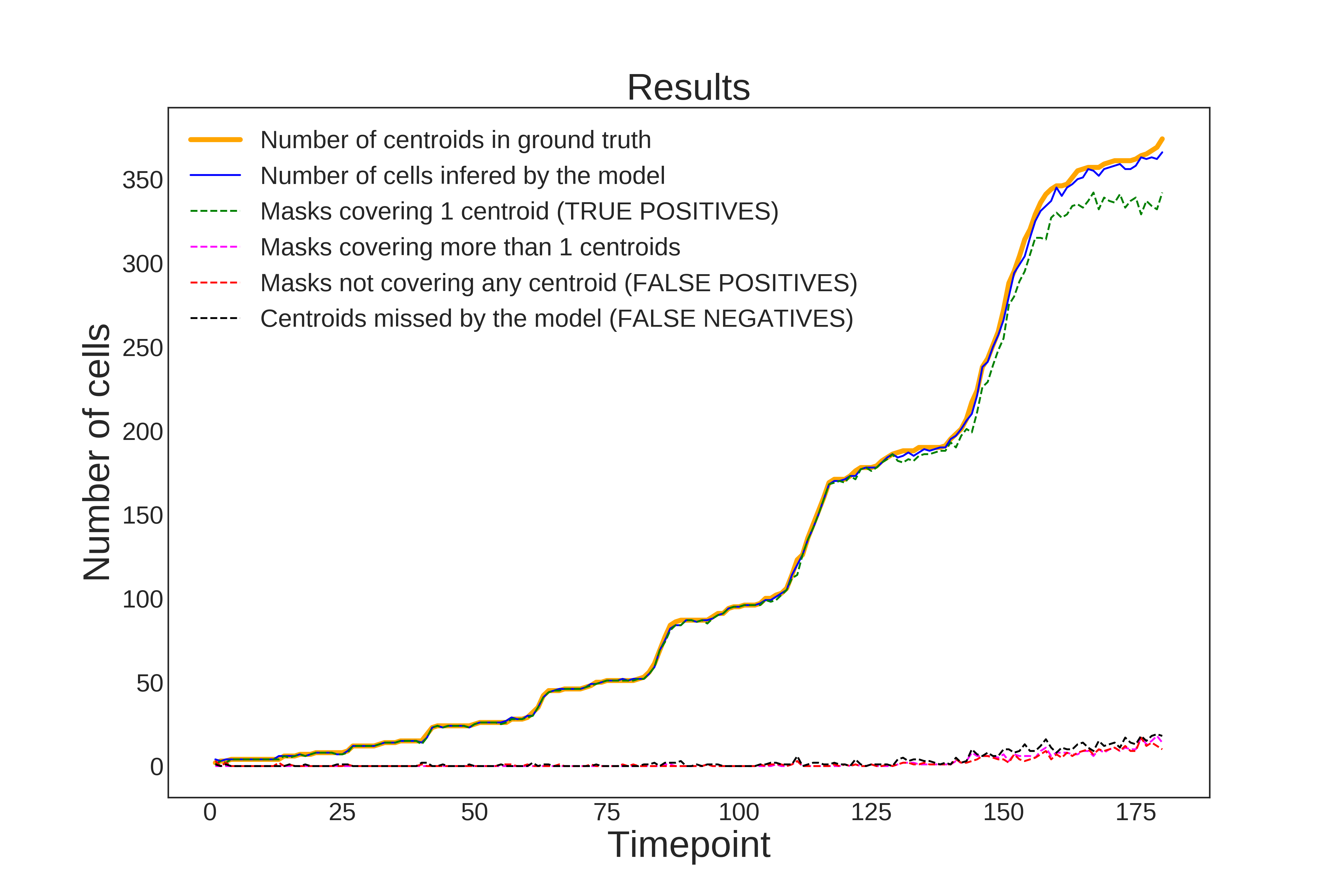 |
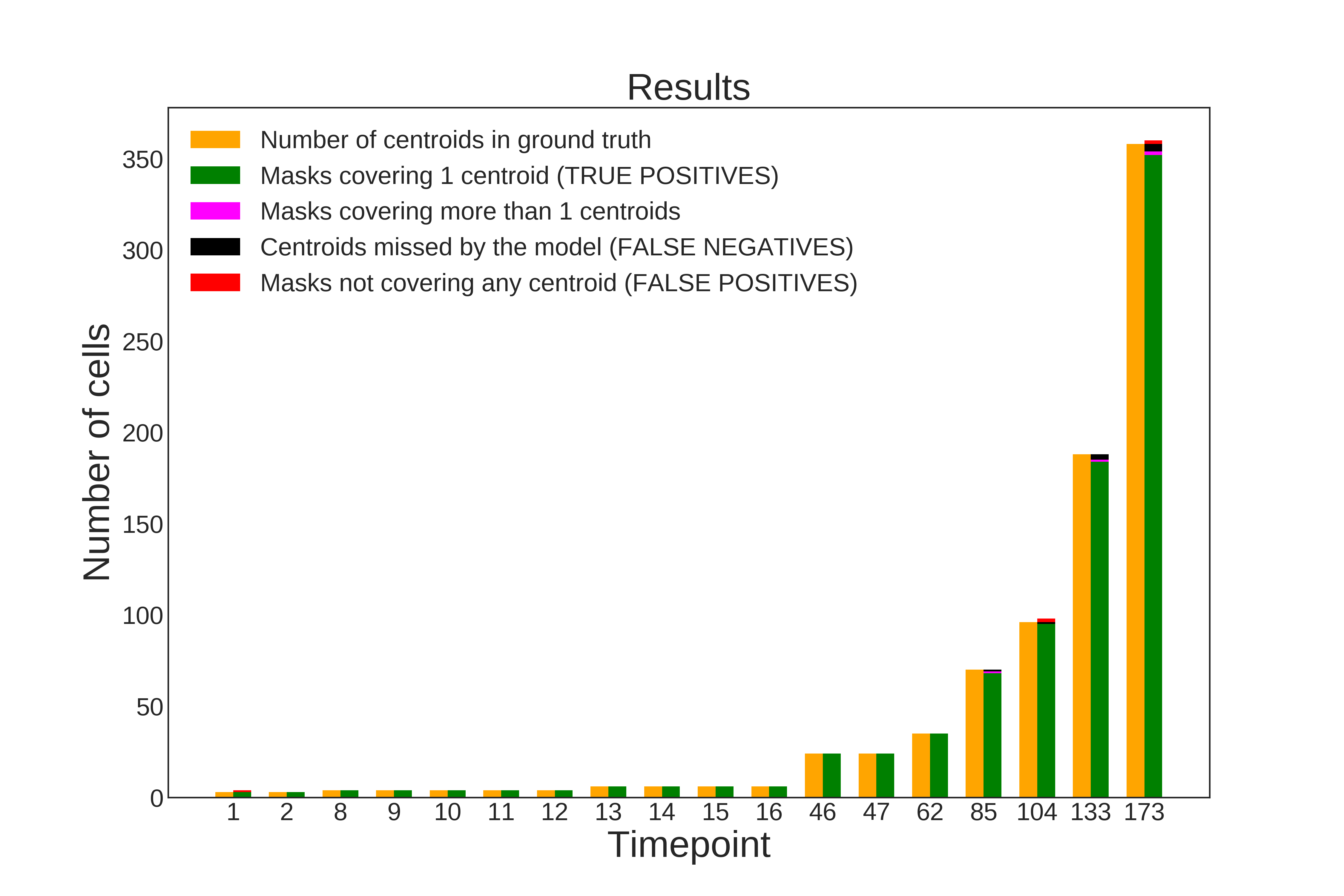 |
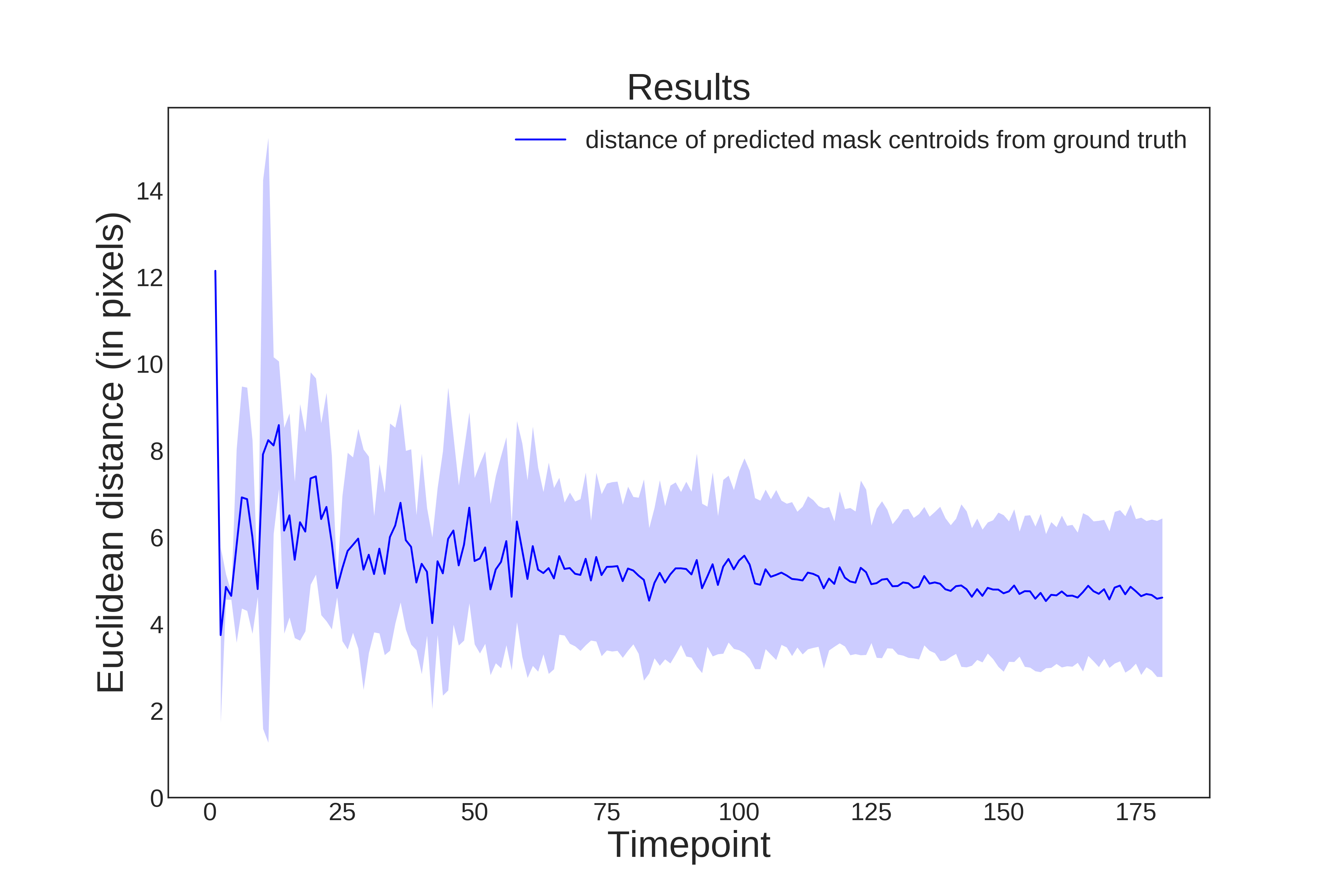 |
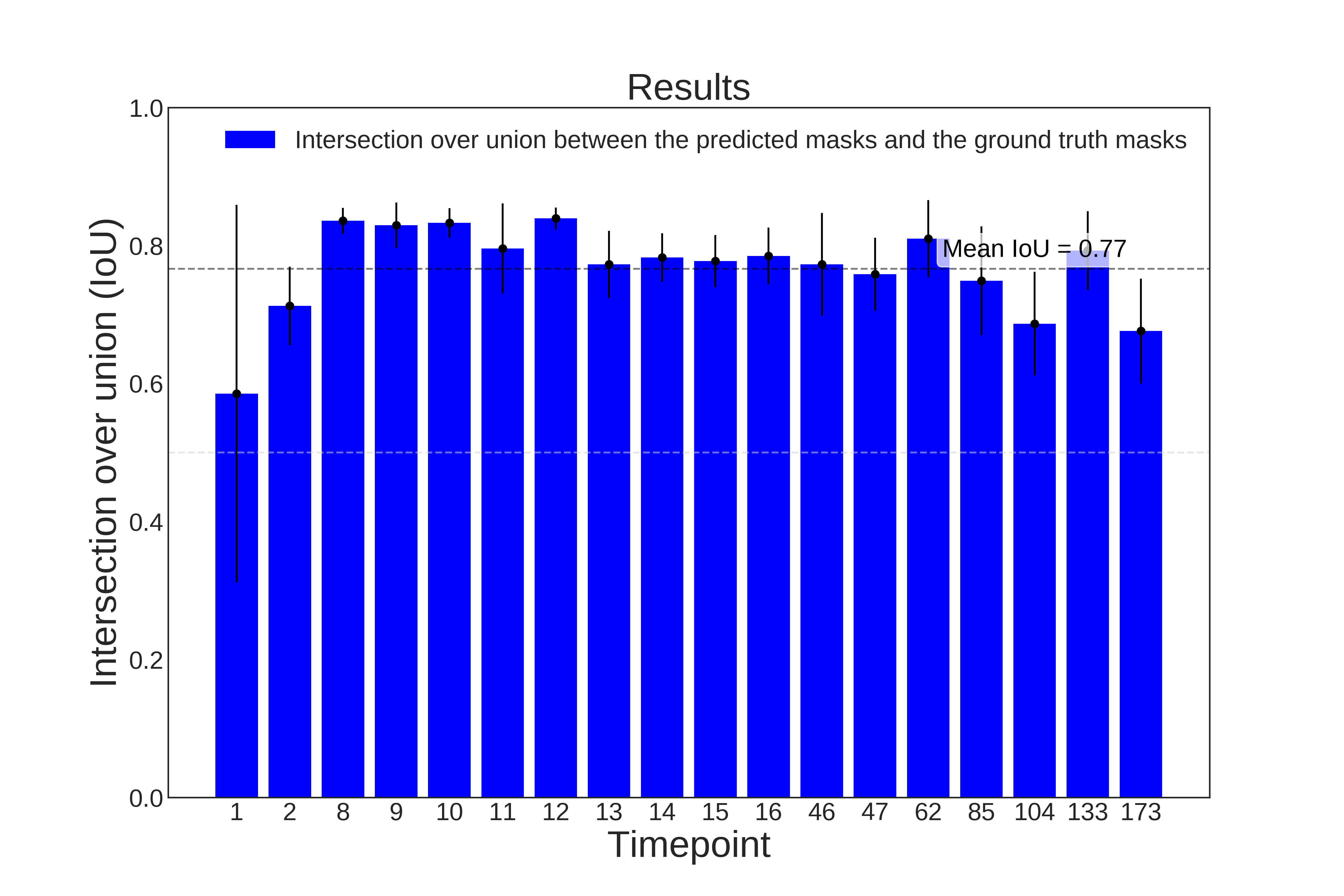 |
You can use the Jupyter Notebook for StarDist3D training from the examples to train a StarDist3D model.
You can use the Jupyter Notebook for StarDist3D predictions from the examples to make predictions using a trained StarDist3D model.
You can use the Jupyter Notebook for StarDist3D evaluations from the examples to evaluate the model's predictions using nuclear centroids or nuclear masks as ground truths.