This is a simple prototypic implementation of a logger that lets you store results on disk in a hierarchical structure (with directories) and retrieve those results into a Pandas dataframe.
The package is based on the assumption that you have a given set of results for a given set of parameters (i.e. that a combination of the paramaters can generate one or more results).
from hierarchical_results import HierarchicalResults, ParameterCombinations
parameters = ["method", "genome", "chromosome"]
results = ["accuracy", "runtime"]
hr = HierarchicalResults(parameters, results)
hr.store_result(["bwa", "hg38", "chr1"], "accuracy", 0.99)
hr.store_result(["bwa", "hg38", "chr1"], "runtime", 100)
hr.store_result(["bwa", "hg38", "chr2"], "accuracy", 0.93)
hr.store_result(["bwa", "hg38", "chr2"], "runtime", 102)
hr.store_result(["minimap", "hg38", "chr1"], "accuracy", 0.97)
hr.store_result(["minimap", "hg38", "chr1"], "runtime", 15)
hr.store_result(["minimap", "hg38", "chr2"], "accuracy", 0.94)
hr.store_result(["minimap", "hg38", "chr2"], "runtime", 16)
# We want to get all results for a given combinations of parameters
# We use the ParameterCombinations class to represent combinations of parameters
combinations = ParameterCombinations(hr.get_names(), [["bwa", "minimap"], "hg38", ["chr1", "chr2"]])
dataframe = hr.get_results_dataframe(combinations, ["accuracy", "runtime"])
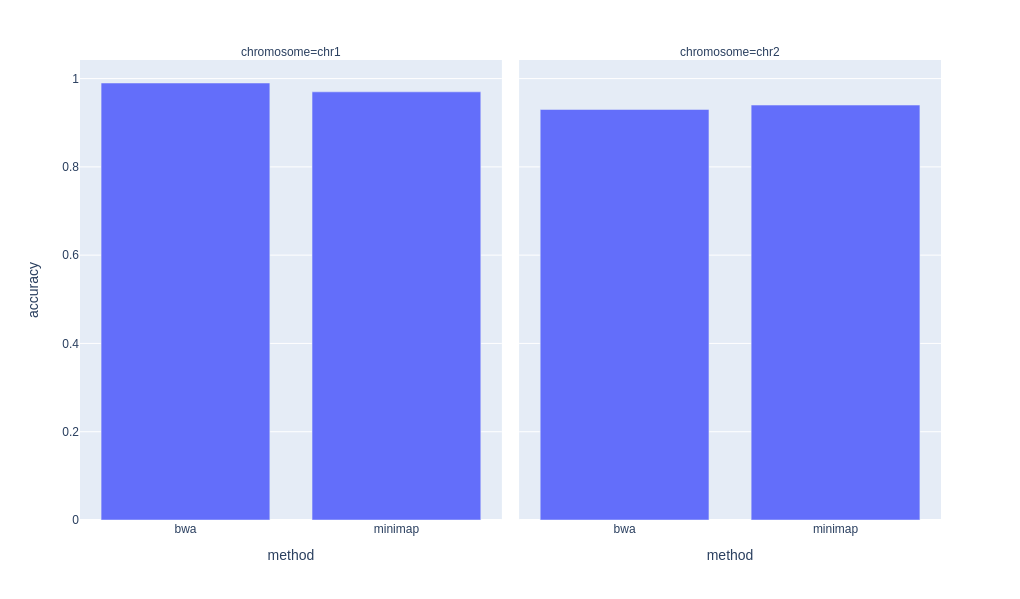
print(dataframe)The above gives a dataframe with the results for the given combinations. You will also have file on disk (e.g. bwa/hg38/chr1/accuracy.txt etc. with the results).
method genome chromosome accuracy runtime
0 bwa hg38 chr1 0.99 100.0
1 bwa hg38 chr2 0.93 102.0
2 minimap hg38 chr1 0.97 15.0
3 minimap hg38 chr2 0.94 16.0
With plotly, we can easily plot this dataframe:
import plotly.express as px
px.bar(dataframe, x="method", y="accuracy", facet_col="chromosome").. which generates this plot:
The HierarchicalResults object can be thought of as a configuration for a "logger". This object can be saved to file. A typical scenario would be to store results while running stuff, then initiating the same object later to gather results and present them.
from shared_memory_wrapper import to_file, from_file
to_file("my_logger", hr)
hr2 = from_file("my_logger")