Automatically tag content tags to taxons using machine learning algorithms.
- Python 3.4.6
- See base-requirements.txt for python dependencies.
- The amazon web services (AWS) command line interface (CLI), see below.
A number of environment variables need to be set before running the cleaning scripts on your system:
| ENV VAR | Description | Nominal value |
|---|---|---|
| DATADIR | Path to the directory storing the data | ./data (relative to the root of the repository -- you may need to set an absolute path) |
| LOGGING_CONFIG | Path to the logging configuration file | ./python/logging.conf (relative to the root of the repository -- you may need to set an absolute path) |
| S3BUCKET | Path of the S3 bucket in which the data are stored. | s3://buod-govuk-taxonomy-supervised-learning |
The Makefile assumes that the python3 command is pointing to the correct distribution of python, which was 3.4.6 in development. To install the correct package dependencies run make pip_install from the project root.
The taxonomy pipeline script runs on the GOV.UK Deploy Jenkins machine:
https://deploy.publishing.service.gov.uk/job/govuk_taxonomy_supervised_learning/
It runs every weekday starting at 2 AM and usually takes a long time to finish.
The content.json.gz and taxon.json.gz files are the raw data files downloaded from the live site and can be downloaded by using scp:
scp deploy.publishing.service.gov.uk:/var/lib/jenkins/workspace/govuk_taxonomy_supervised_learning/data/* .
These files need to be moved to DATADIR
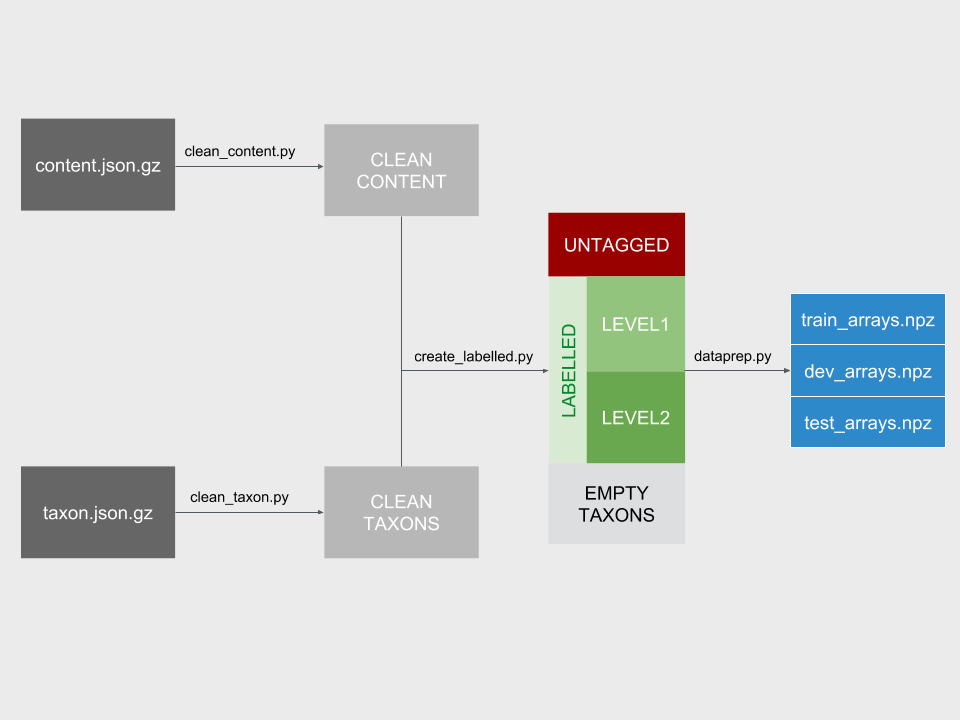
After setting environment variables and obtaining the raw data files saved in DATADIR, running make will download the data and launch the cleaning scripts in order. The following files are created by the various cleaning scripts:
| source filename (data/) | output filename (data/) | produced by (python/) |
|---|---|---|
| taxons.json.gz | clean_taxons.csv.gz | clean_taxons.py |
| content.json.gz | clean_content.csv | clean_content.py |
| clean_taxons.csv.gz; clean_content.csv; content_to_taxon_map.csv | untagged.csv.gz | create_labelled.py |
| clean_taxons.csv.gz; clean_content.csv; content_to_taxon_map.csv | empty_taxons.csv.gz | create_labelled.py |
| clean_taxons.csv.gz; clean_content.csv; content_to_taxon_map.csv | labelled.csv.gz | create_labelled.py |
| clean_taxons.csv.gz; clean_content.csv; content_to_taxon_map.csv | labelled_level1.csv.gz | create_labelled.py |
| clean_taxons.csv.gz; clean_content.csv; content_to_taxon_map.csv | labelled_level2.csv.gz | create_labelled.py |
| labelled*.csv.gz | *arrays.npz | dataprep.py |
The following schematic describes the movement of data through the pipeline, and the role of each of the scripts.
The cleaned files are used by the python notebooks contained in python/notebooks.
You should use your virtualenv when running jupyter notebooks. Follow the following steps:
Install the ipython kernel module into your virtualenv
workon my-virtualenv-name # activate your virtualenv, if you haven't already
pip install ipykernel
Now run the kernel "self-install" script:
python -m ipykernel install --user --name=my-virtualenv-name
Replacing the --name parameter as appropriate.
You should now be able to see your kernel in the IPython notebook menu: Kernel -> Change kernel and be able so switch to it (you may need to refresh the page before it appears in the list). IPython will remember which kernel to use for that notebook from then on.
| Name | Activity | Data inputs | Data outputs |
|---|---|---|---|
| EDA-count-data | Read in and count data files | untagged_content.csv, clean_taxons.csv, clean_content.csv.gz, labelled.csv, filtered.csv, empty_taxons.csv, old_tags.csv | None |
| EDA-taxons | Descriptive analysis of taxon content overall, and according to level | labelled, filtered, taxons | level2taxons_concordant.csv, taggedtomorethan10taxons.csv |
| EDA-document-type | Descriptive analysis of content according to document type, over time | untagged, labelled, filtered, labelled_level1, labelled_level2 | document_type_group_lookup.json |
| EDA-other-metadata | Descriptive analysis of content according to metadata types, over time | untagged, labelled, filtered, labelled_level1, labelled_level2 | none |
| Name | Activity | Data inputs |
|---|---|---|
| CNN-allgovuk.ipynb | Convolutional Neural Network of tagged content using keras framework and pre-trained word embeddings | clean_content.csv.gz, clean_taxons.csv |
| SVM_allgovuk.ipynb | Support vector machine of tagged content | |
| TPOT_allgovuk.ipynb | Genetic algorithm to select optimal algorithm and hyperparameters |
| Name | Activity | Data inputs | Data outputs |
|---|---|---|---|
| EDA | Exploratory data analysis | untagged_content.csv, clean_taxons.csv, clean_content.csv.gz | None |
| clean_content.ipynb | Development of steps to process raw content data into formats for use in EDA and modelling. These are now used in clean_content.py, which is called by the Makefile | ||
| explore_content_dupes.ipynb | Understand duplicates in gov.uk content items | raw_content.json, clean_content.csv | None |
The default logging configuration used by the data transformation pipeline (set in ./python/) will do the following things:
- Write a simple log to stdout (console) at
INFOlevel - Write a more detailed log to a file at
DEBUGlevel (by default/tmp/govuk-taxonomy-supervised-learning.log).
Setting up GPU-backed instances on AWS is greatly facilitated by using databox. Currently the features required to create a deep learning instances are in Pull Request 31. Once these are merged into master, operate databox from master, but for now you will need to git checkout feature/playbook_argument. Once you have databox and all its dependencies installed, the following command will instantiate an instance prepared for Deep Learning on AWS:
./databox.sh -a ami-1812bb61 -r eu-west-1 -i p2.xlarge -s snap-04eb15f2e4faee97a -p playbooks/govuk-taxonomy-supervised-learning.yml up
The arguments are explained in the table below:
| Argument | Value | Description |
|---|---|---|
| -a | ami-1812bb61 | The Conda based Amazon Machine Image. Other options are explained by amazon. |
| -i | p2.xlarge | This is the smallest of the Deep Learning instance types. More information is available here. Note that the Deep Learning AMIs may not work with the newer p3 GPU instances. |
| -s | snap-04eb15f2e4faee97a | The id of the snapshot containing the taxonomy data. This can be checked at the AWS console |
| -p | playbooks/govuk-taxonomy-supervised-learning.yml | The ansible playbook describing deployment tasks required on to setup the instance. |
| -r | eu-west-1 | The region in which the instance will be deployed. At present this must be set to eu-west-1 (Ireland) as some deep learning instances are not available in the eu-west-2 (London) zone, and the snapshot is currently in eu-west-1 (although could be copied elsewhere. |
Once the instance has instantiated, you will need to run the following commands:
- SSH tunnel into the instance with
ssh -L localhost:8888:localhost:8888 ubuntu@$(terraform output ec2_ip) - Open tmux to ensure that any operations do not fail if you disconnect
- Activate the tensorflow_p36 environment and run
jupyter notebookon the instance:
tmux
source activate tensorflow_p36
jupyter notebook
This will set up a notebook server, for which you will be provided a link in the console.
- Log in to notebook server on your local machine by copying the link generated on the server into a browser. This will give you access to jupyter notebooks on your local machine.
- To run tensorboard ensure that the tensorboard callback has been enabled in the model, then log into the instance again in a new terminal creating a new tunnel with
ssh -L localhost:6006:localhost:6006 ubuntu@$(terraform output ec2_ip). - Open tmux to ensure the task continues running even if you disconnect.
- Activate the
tensorflow_p36environment withsource activate tensorflow_p36. - Run
tensorboard --log_dir=<path to logging>. - Open a browser on your local machine and navigate to https://localhost:6006 to access the tensorboard sever.
- Ensure that your model is running on the instance GPU by running
nvidia-smiin a new terminal on the instance (you can run this repeatedly withwatch -n 10 nvidia-smito update every 10 seconds).