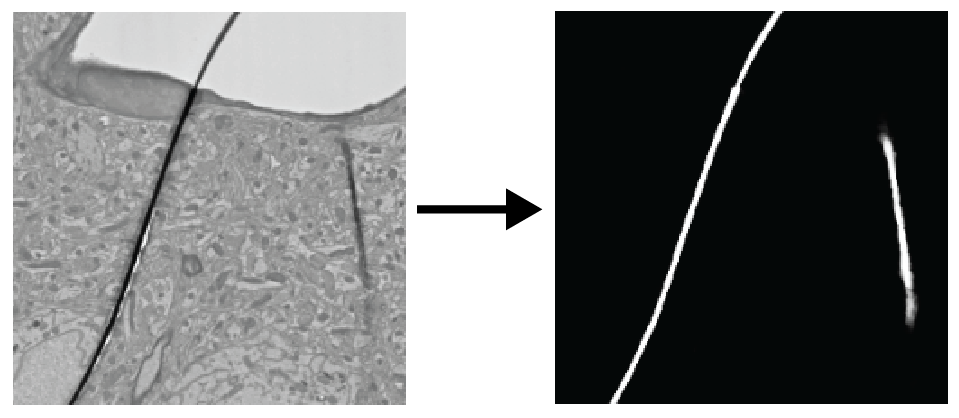
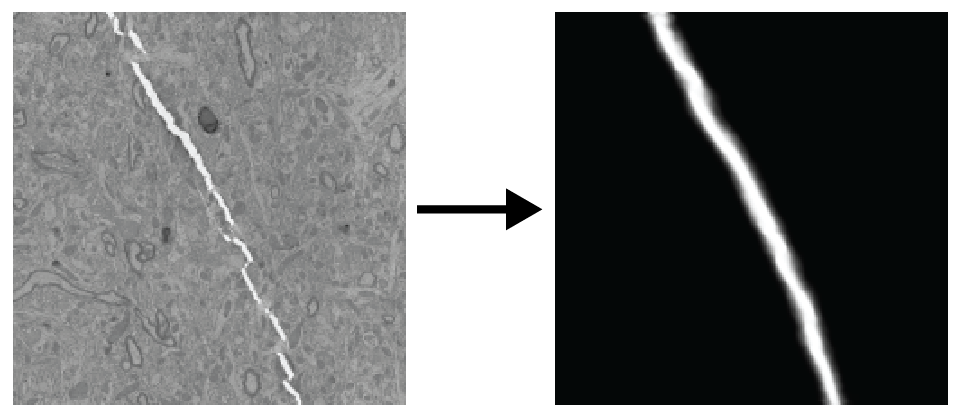
Deep learning to detect particular features on EM images.
- Defect (fold/crack) detection
- Fold
- Crack
- Resin detection
- Film detection
- Mitochondria detection
conda create -n cmito python=3.8
git clone https://github.com/jabae/Cmito.git
cd Cmito
pip install -r requirements.txt
python train.py --exp_dir /experiment_directory/
--train_image /train_image.h5 --train_label /train_mask.h5
--val_image /val_image.h5 --val_label /val_mask.h5 --chkpt_num 0
python inference.py --exp_dir /experiment_directory/ --chkpt_num 150000
--input_file test_image.h5 --output_file pred_mask.h5
Large-scale inference can be done using the SEAMLeSS module.
Use alex-emdetector branch.
- Single-label inference
python3 run_emdetector.py --model_path ../models/Zetta_RFNet0120/
--src_path gs://zetta_aibs_human_unaligned/mip3_stack --dst_path gs://zetta_aibs_human_unaligned/masks/resin/resin_prelim
--bbox_start 0 0 5100 --bbox_stop 1048576 1048576 5102
--bbox_mip 0 --max_mip 8 --mip 6 --chunk_size 2048 2048 --overlap 32 32
- Multi-label inference
python3 run_multi_emdetector.py --model_path ../models/Zetta_RFNet0120/
--src_path gs://zetta_aibs_human_unaligned/mip3_stack --dst1_path gs://zetta_aibs_human_unaligned/masks/resin/resin_prelim
--dst2_path gs://zetta_aibs_human_unaligned/masks/film/film_prelim --bbox_start 0 0 5100 --bbox_stop 1048576 1048576 5102
--bbox_mip 0 --max_mip 8 --mip 6 --chunk_size 2048 2048 --overlap 32 32
Parts of the codes have been adopted from DeepEM.