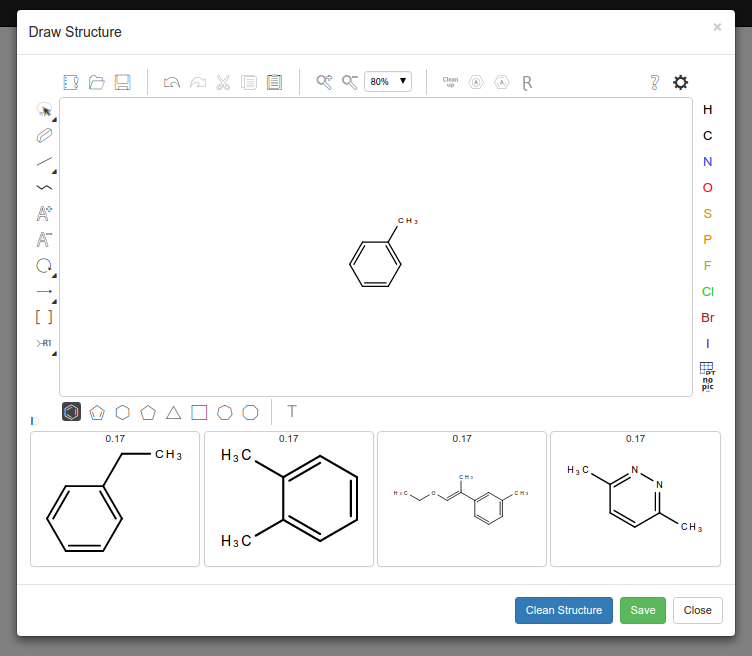
This repo contains code to create a molecular strucutre graph wihtin an SQL database. While the user is drawing it takes the data and creates a baysin network to try and predict what the user is trying to create from past data.
Move into the /rest-api sub-directory
- Clone RDKit repo using command:
gradle cloneRDKit`
- you are required to generate the RDkit wrappers:
buildRDKit.sh`
- Build overall project:
gradle build
Move into the /front-end sub-directory
-
run npm install
-
Apply for getting the Ketcher 2.0 code from:
http://lifescience.opensource.epam.com/download/ketcher.html
- Place the ketcher lib into directory /front-end/lib/
- You now need to download and run hsqldb.
-
Take database.script file from /rest-api folder.
-
run with command from hsqldb folder:
sudo java -cp hsqldb.jar org.hsqldb.Server -database.0 file:database.script -dbname.0 jacr_structure_prediction
- set /rest-api/application.properties
spring.datasource.url=jdbc:hsqldb:hsql://localhost:9001/jacr_structure_prediction
bash /rest-api/run.sh
open file
front-end/struct-predicion.html