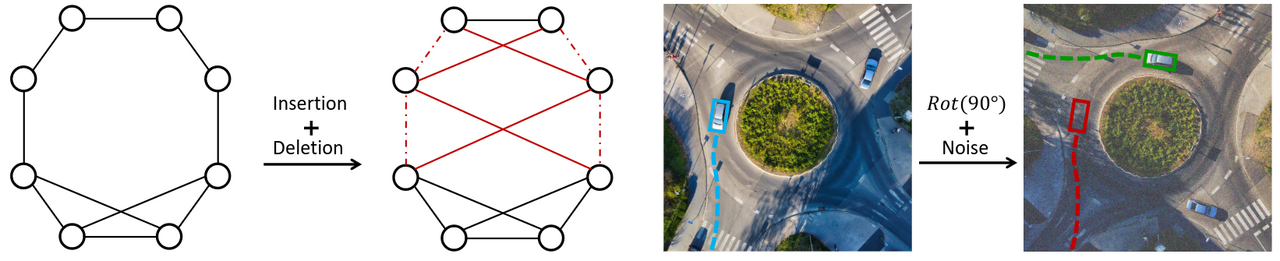
This is the official implementation of
"Provable Adversarial Robustness for Group Equivariant Tasks: Graphs, Point clouds, Molecules, and More"
Jan Schuchardt, Yan Scholten, and Stephan Günnemann, NeurIPS 2023.
To install the requirements, execute
conda env create -f environment.yml
You also need to download reference implementations of different geometric machine learning models and certificates, which we extend.
They can be downloaded to the reference_implementations folder via
git submodule init
git submodule update
You then need to install each of the packages in the reference_implementations folder.
This can usually be done via pip install -e . and/or conda install --name equivariance_robustness --file [...].yml. Consult their respective readmes.
To train molecular force prediction models via the code in reference_implementations/uncertainty_molecules, you will additionally need to create and configure a Weights & Biases account.
You can install this package via pip install -e .
The graph datasets (TUDataset and Planetoid) are downloaded automatically via pytorch geometric.
The molecule dataset (MD17) is also downloaded automatically via pytorch geometric.
For the ModelNet40 dataset, download the original and pre-processed files linked in the Pointnet_Pointnet2_pytorch repository.
In order to reproduce all experiments, you will need need to execute the scripts in seml/scripts using the config files provided in seml/configs.
We use the SLURM Experiment Management Library, but the scripts are just standard sacred experiments that can also be run without a MongoDB and SLURM installation.
After computing all certificates, you can use the notebooks in plotting to recreate the figures from the paper.
In case you do not want to run all experiments yourself, you can just run the notebooks while keeping the flag overwrite=False (our results are then loaded from the respective raw_data files).
For more details on which config files and plotting notebooks to use for recreating which figure from the paper, please consult REPROCE.MD.
Please cite our paper if you use this code in your own work:
@InProceedings{Schuchardt2023_Equivariance,
author = {Schuchardt, Jan and Scholten, Yan and G{\"u}nnemann, Stephan},
title = {Provable Adversarial Robustness for Group Equivariant Tasks: Graphs, Point Clouds, Molecules, and More},
booktitle = {Conference on Neural Information Processing Systems (NeurIPS)},
year = {2023}
}