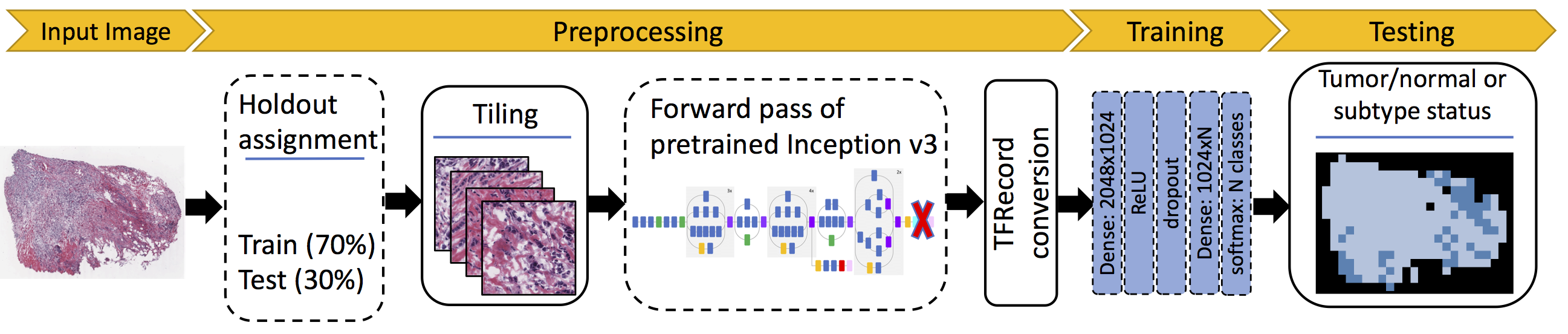
This package is an implementation of Inception-v3 CNN to classify TCGA H&E whole slide images (WSI) according to tumor/normal status and cancer subtype. It implements a pipeline in Kubernetes (k8s) under Google Cloud Platform (GCP) for labeling, tiling, and transfer learning on the images.
For details, please see the following paper:
Javad Noorbakhsh, Saman Farahmand, Sandeep Namburi, Dennis Caruana, David Rimm, Mohammad Soltanieh-ha, Kourosh Zarringhalam, Jeffrey H Chuang, Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images, Nature communications 2020
To properly use the pipelines, you will need to install the package. After changing current directory to the root of the project run (for now this only works in the development mode):
pip install -e .The Kubernetes pipelines are separated into individual apps which run the corresponding problem of interest on GCP. For details on each app refer to the README in its corresponding folder. The following apps are implemented:
- k8s-app-tile: tile whole slide images
- k8s-app-createcaches: apply a forward pass of tiles through pretraind CNN and store the last layer values as a text file (hereafter called 'cache')
- k8s-app-cache-tfrecords-tn: create tfrecords from caches for tumor/normal classification
- k8s-app-cache-tfrecords-subtype: create tfrecords from caches for subtype classification
- k8s-app-runcnn-tn: run CNN for tumor/normal classification
- k8s-app-runcnn-subtype: run CNN for subtype classification
- k8s-app-runcnn-tn-crossclassify: test the CNNs trained on tumor/normal status for cross-classification
To begin using any of these apps you will need to set up a k8s cluster.
Few functionalities have been implemented through a command line tool. To access its help run:
histcnn --helpor one of the more detailed alternatives:
histcnn gcs --help
histcnn annotate --help
histcnn run-subtype --help