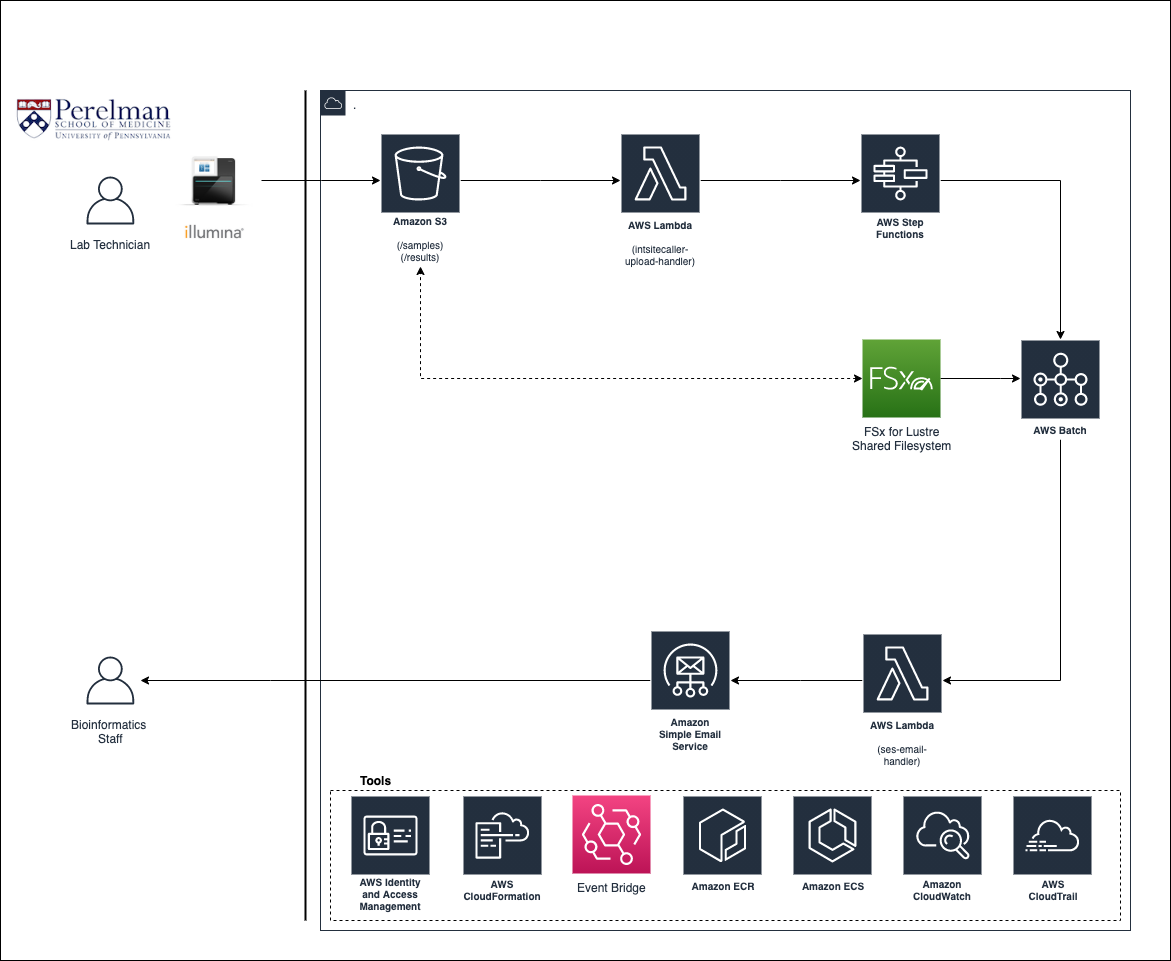
This project contains CloudFormation templates to configure an AWS Batch-based genomics workflow.
You must have the aws cli installed and configured to connect to your account.
Follow this link for installation directions:
https://docs.aws.amazon.com/cli/latest/userguide/install-cliv2.html
You'll want to run aws configure to set up a profile to connect to your AWS account.
More on that here:
https://docs.aws.amazon.com/cli/latest/userguide/cli-chap-configure.html
You will need to configure SES in your account, if you want to use the SES email lambda handler part of the solution, found in Alerts.yml
Details here:
https://docs.aws.amazon.com/ses/latest/DeveloperGuide/send-email-set-up.html
See create_from_scratch.sh for detailed instructions on how to create the environment.
- You will need to update To and From email addresses in the Alerts.yml file.
The templates contained in this project will create the architecture depicted below, which is based off of the Secondary Analysis reference architecture described in the AWS Genomics Whitepaper, available here: