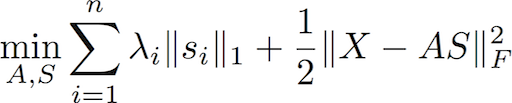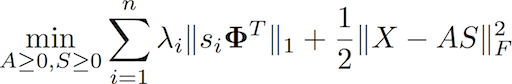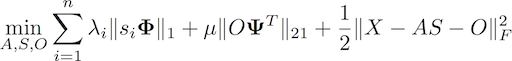Toolbox for solving sparse matrix factorization problems
This toolbox is composed of the following submodules:
-
GMCA is the building block of the pyGMCALab toolbox. This algorithm basically tackles sparse BSS problems :
-
Building upon GMCA, AMCA is an extension that specifically deals with partially correlated sources
-
nGMCA allows solving sparse non-negative matrix factorization problems (sparse NMF). Sparse modelling can be done either in the sample domain or in a transformed domain. For that purpose, a python/C wrapper for wavelets (RedWave) is also provided as an external module (see ./redwave_toolbox).
-
rGMCA copes with sparse BSS problems in the presence of outliers. Tr_rGMCA is an extension that can benefit from the morphological diversity between the outliers and the sources.
For all these algorithms, we strongly advise the interested user to have a close look at the jupyter notebooks, which are provided in ./pyGMCA/scripts
It tackles sparse blind source separation (BSS) problems of the form:
One of the aspects of the GMCA algorithm is that the regularization parameters are automatically tuned based on the noise level. The latter is estimated straight from the data thanks to an empirical estimator coined the Median Absolute Deviation (MAD). The current code assumes that the data are already expressed in the sparse domain. A first step then consist in applying your favorite sparsifying transform to the input data X prior to perform the GMCA algorithm.
One of the main limitations of most sparse BSS methods is that they rely on separation principles such as statistical independence for ICA-based methods or morphological component analysis for GMCA, which rarely holds in real-world applications. In many applications, the sources of interest generally exhibit some partial correlations that are not correctly accounted for by classical approaches. For that purpose, an extension of the GMCA algorithm has been introduced in [1], which allows accounting for partial correlations. Both GMCA and AMCA can be applied using the same basic code, with the exception of a single change of option value.
For more details about the GMCA algorithm, we refer the interested reader to:
- J.Bobin, J.-L. Starck, Y.Moudden, J. Fadili, Blind Source Separation: the Sparsity Revolution
- J.Bobin , J. Rapin, J.L. Starck and A. Larue, Sparsity and adaptivity for the blind separation of partially correlated sources
It tackles sparse non-negative matrix factorization problems (NMF) problems of the form:
where Φ stands for the sparse representation. One novelty of the nGMCA algorithm is that it makes use of recent solvers for non-smooth convex optimization problems such as the (Generalized) Forward Backward splitting algorithm (FBS). The pyGMCALab toolbox provides implementations of the FBS to tackle the basic subproblems that compose the nGMCA algorithm.
For more details, we refer the interested user to:
- J.Rapin, J.Bobin, A. Larue and J.L. Starck, Sparse and non-negative BSS for noisy data
- J. Rapin, J.Bobin, A. Larue and J.L. Starck, NMF with sparse regularizations in transformed domains
It tackles sparse robust BSS problems of the form:
where the term O stands for sparse outliers. For more details, we refer the interested user to:
- Undecimated wavelets with the pyredwave toolbox
The algorithms using sparsity in a transformed domain need the pyredwave Toolbox: a specific toolbox computing 1D or 2D wavelet transform on any 1 or 2 dimensions of an up to 4 dimensional data. Execute ”python setup.py build” in a terminal from the pyredwave folder so as to build it. The compilation requires Boost.Python (tested on Mac and Ubuntu, with Python 2.7). The toolbox uses OMP for CPU parallelization. To disable parallelization, remove the tag ” PARALLELIZED ” in pyredwave/pyredwave/cxx/redWaveTools.hpp
- cxxStarlet
The folder cxxStarlet contains C/python wrappers to compute the starlet transform (tested on Mac and Ubuntu, with Python 2.7). The toolbox uses OMP for CPU parallelization. compilation is done as follows:
- Edit the CMakeList file and comment the desired command line (For MAC or For Linux) depending on the computer type
- Go to the build subfolder and type cmake .. (please note that cmake is required)
- If some libraries are missing (e.g. gsl, cfitsio, boost), you'll be notified to set them up
- Type make, this will yield a sparse2d.so file that you will be able to import in python
Contributors include: J.Bobin, C.Chenot, C.Kervazo, J.Rapin. Part of this work was funded European Community through the grant LENA (ERC StG no. 678282) within the H2020 Framework Program.