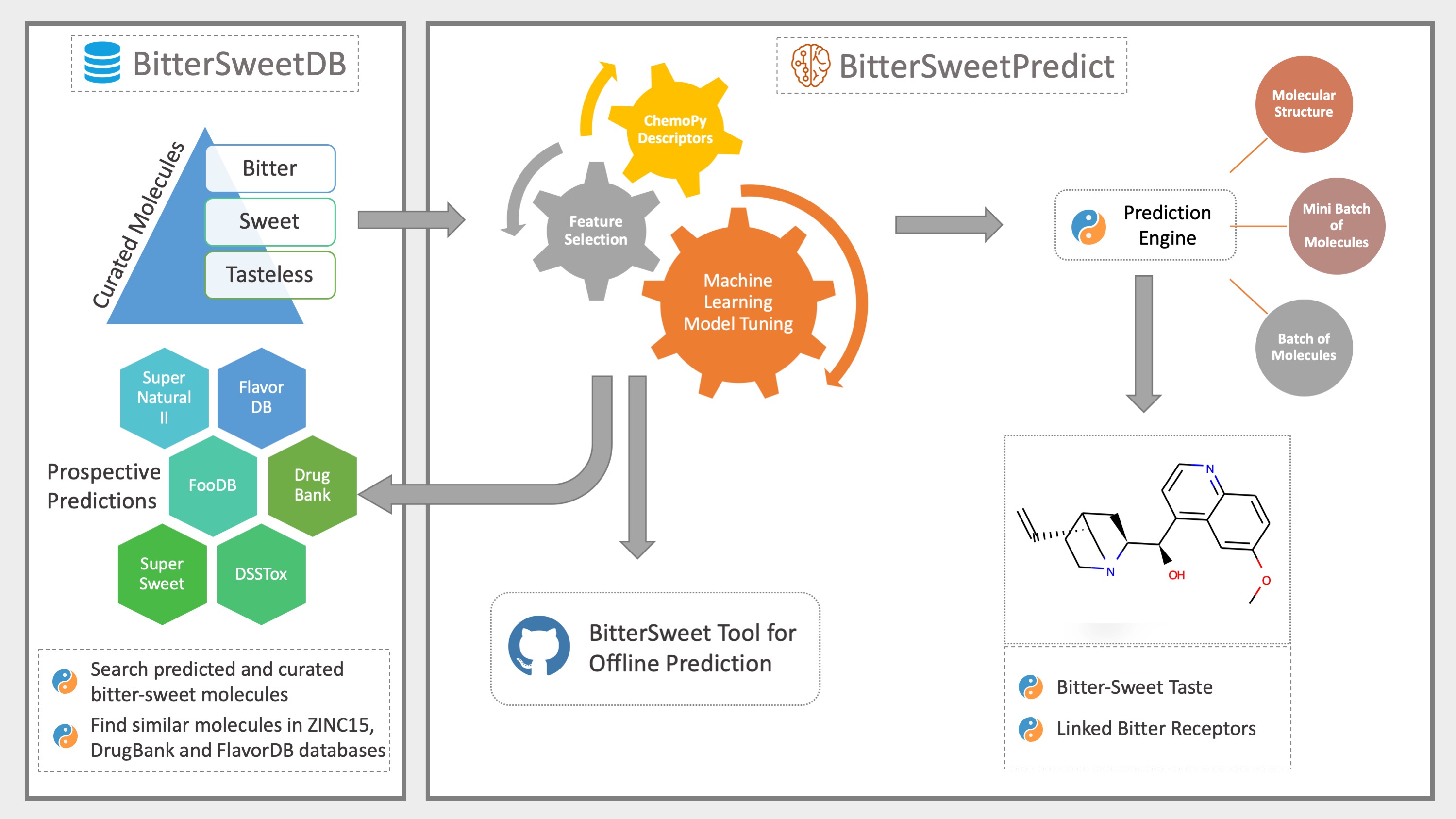
BitterSweet: Building machine learning models for predicting the bitter and sweet taste of small molecules
The dichotomy of sweet and bitter tastes is a salient evolutionary feature of human gustatory system with an innate attraction to sweet taste and aversion to bitterness. A better understanding of molecular correlates of bitter-sweet taste gradient is crucial for identification of natural as well as synthetic compounds of desirable taste on this axis. While previous studies have advanced our understanding of the molecular basis of bitter-sweet taste and contributed models for their identification, there is ample scope to enhance these models by meticulous compilation of bitter- sweet molecules and utilization of a wide spectrum of molecular descriptors. Towards these goals, based on structured data compilation our study provides an integrative framework with state-of-the-art machine learning models for bitter-sweet taste prediction (BitterSweet). We compare different sets of molecular descriptors for their predictive performance and further identify important features as well as feature blocks. The utility of BitterSweet models is demonstrated by taste prediction on large specialized chemical sets such as FlavorDB, FooDB, SuperSweet, Super Natural II, DSSTox, and DrugBank. To facilitate future research in this direction, we make all datasets and BitterSweet models publicly available, and also present an end-to-end software for bitter-sweet taste prediction based on freely available chemical descriptors.
- Rudraksh Tuwani
- Somin Wadhwa
- Ganesh Bagler*
Center for Computational Biology, Indraprastha Institute of Information Technology (IIIT- Delhi), New Delhi, India *Corresponding Author (ganesh.bagler@gmail.com, bagler@iiitd.ac.in)
To setup a working environment to execute some or all sections of this project, you must:
-
Clone the project
bittersweet-$ git clone https://github.com/cosylabiiit/bittersweet.git $ cd bittersweet -
We use
condaas a tool to create isolated virtual environments and since some of our packages require building binaries from their source, it is necessary to create your env from therequirement.ymlfile provided.$ conda env create -f environment.yml $ conda activate env -
To deactivate this environment after usage -
$ conda deactivate
* Ensure that all scripts are run under a python 2.7 environment.
.
.
├── data # Model Training & Test Data (Tabular Format)
│ ├── bitter-test.tsv
│ ├── bitter-train.tsv
│ ├── sweet-test.tsv
│ ├── sweet-train.tsv
├── bittersweet # All Source Files
│ ├── models # Trained Models
│ │ ├── bitter_chemopy_boruta_features.p
│ │ ├── bitter_chemopy_rf_boruta.p
│ │ ├── sweet_chemopy_boruta_features.p
│ │ ├── sweet_chemopy_rf_boruta.p
│ ├── __init__.py
│ ├── model.py
│ ├── properties.py
│ ├── read_file.py
├── manuscript-experiments # Testing modules (including those for random-control experiments)
│ ├── bittersweet # Directory containing scripts
│ ├── data # Directory containing data
│ ├── models # Directory containing models
├── examples
├── predict.py # methods to test our models
.
.
The authors thank Indraprastha Institute of Information Technology (IIIT-Delhi) for providing computational facilities and support.
G.B. and R.T. designed the study. R.T. curated the data. S.W., R.T. performed feature selection and importance ranking experiments, and trained the models. R.T. generated the bitter-sweet predictions for specialized chemicals sets. All the authors analysed the results and wrote the manuscript.