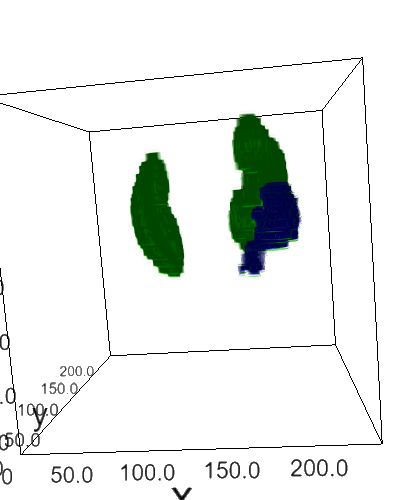
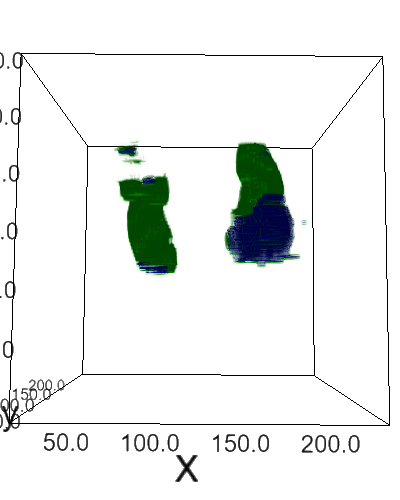
Using 2D & 3D convolutional neural networks for the 2019 Kidney and Kidney Tumor Segmentation Challenge. This repository is associated with this conference paper (older version).
I'm not sure why the tumor scores are so low for all of the architectures, so I'm open to any suggestions and PRs! Am actively working on improving them.
Major credits to:
- Isensee's nnU-Net and batchgenerators
- qubvel's segmentation_models.pytorch
- Nick Heller for hosting KiTS19!
- Resampling to 3.22 × 1.62 × 1.62 mm
- Isensee's nnU-Net methodology
- Clipping to the [0.5, 99.5] percentiles and applying z-score standardization
- Foreground class sampling
- 2D: Done by sampling per slice (loading only 2D arrays)
- SO MUCH FASTER THAN LOADING 3D ARRAYS
- Difference: 3 seconds v. 5 minutes per epoch
- SO MUCH FASTER THAN LOADING 3D ARRAYS
- 3D: Done through
ROICropTransform
- 2D: Done by sampling per slice (loading only 2D arrays)
- Data Augmentation
- Located in
kits19cnn/experiments/utils.py- Pay attention to the
augmentation_keys inget_training_augmentationandget_validation_augmentation
- Pay attention to the
- Done through
batchgenerators+ my own custom transforms
- Located in
- SGD (lr=1e-4) and LRPlateau (factor=0.15 and patience=5); BCEDiceLoss
- 2D: batch size = 18 (regular training)
- 3D: batch size = 4 (fp16 training)
- 2D (patch size: (256, 256))
- Vanilla 2D nnU-Net
- 6 pools with convolutional downsampling and upsampling
- max number of filters set to 320 and the starting number is 30
- 2D U-Net with pretrained ImageNet classifiers
- 2D FPN with pretrained ImageNet classifiers
- Vanilla 2D nnU-Net
- 3D (patch size: (96, 160, 160))
- 3D nnU-Net
- 5 pools with convolutional downsampling and upsampling
- max number of filters set to 320 and the starting number is 30
- 3D nnU-Net (Classification + Segmentation)
- 3D nnU-Net
| Neural Network | Parameters | Local Test (Tumor-Kidney) Dice | Local Test (Tumor Only) Dice | Weights |
|---|---|---|---|---|
| 2D nnU-Net | 12M | 0.90 | 0.26 | ... |
| 3D nnU-Net | 29.6M | 0.86 | 0.22 | ... |
| ResNet34 + U-Net Decoder | 24M | 0.90 | 0.29 | ... |
| ResNet34 + FPN Decoder | 22M | 0.83 | 0.29 | ... |
The recommended way is to just follow the instructions on the original kits19 Github challenge page, which utilizes git lfs.
Here is a brief run-down for Google Colaboratory:
! sudo add-apt-repository ppa:git-core/ppa
! curl -s https://packagecloud.io/install/repositories/github/git-lfs/script.deb.sh | sudo bash
! sudo apt-get install git-lfs
! git lfs install
% cd "/content/"
! rm -r kits19
! git clone https://github.com/neheller/kits19.git
# takes roughly 11 minutes to download
To do general preprocessing (resampling):
# preprocessing
from kits19cnn.io.preprocess import Preprocessor
base_dir = "/content/kits19/data"
out_dir = "/content/kits_preprocessed"
preprocess = Preprocessor(base_dir, out_dir)
preprocess.cases = sorted(preprocess.cases)[:210]
%time preprocess.gen_data()
Note that the standardization and clipping is done on-the-fly.
If you want to do 2D segmentation:
# preprocessing
from kits19cnn.io.preprocess import Preprocessor
out_dir = "/content/kits_preprocessed"
preprocess = Preprocessor(out_dir, out_dir, with_mask=True)
preprocess.cases = sorted(preprocess.cases)[:210]
preprocess.save_dir_as_2d()
If you want to do binary 2D segmentation (kidney only or renal tumor only).
import os
from kits19cnn.experiments.utils import parse_fg_slice_dict_single_class
preprocessed_dir = "/content/kits_preprocessed"
json_path = os.path.join(preprocessed_dir, "slice_indices.json")
out_path = os.path.join(preprocessed_dir, "slice_indices_tu_only.json")
_ = parse_fg_slice_dict_single_class(json_path, out_path, removed_fg_idx="1")
out_path = os.path.join(preprocessed_dir, "slice_indices_kidney_only.json")
_ = parse_fg_slice_dict_single_class(json_path, out_path, removed_fg_idx="2")
Please see the example yaml file at script_configs/train.yml. Works for 2D, 2.5D,
and 3D. Also, supports binary 2D segmentation if you change the slice_indices_path.
Also, supports classification + segmentation for nnU-Net (doesn't work that well).
python /content/kits19-cnn/scripts/train_yaml.py --yml_path="/content/kits19-cnn/script_configs/train.yml"
TensorBoard: Catalyst automatically supports tensorboard logging, so just run this in Colaboratory:
# Load the TensorBoard notebook extension
%load_ext tensorboard
# Run this before training
%tensorboard --logdir logs
For Plotting Support (plotly/orca) [OPTIONAL]:
The regular training script (script_configs/train.yml) doesn't plot the graphs
directly, but saves them as .png files. If you don't want to do all of this installing, just exclude plot_params in scripts/train_yaml.py
# on colab
# installing anaconda and plotly with orca + dependencies
!wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
!chmod +x Miniconda3-latest-Linux-x86_64.sh
!bash ./Miniconda3-latest-Linux-x86_64.sh -b -f -p /usr/local
# !conda install -c plotly plotly-orca
!conda install -c plotly plotly-orca psutil requests ipykernel
!export PYTHONPATH="${PYTHONPATH}:/usr/local/lib/python3.7/site-packages/"
!pip install nbformat
# orca with xvfb support (so orca can save the graphs)
# Plotly depedencies
!apt-get install -y --no-install-recommends \
wget \
xvfb \
libgtk2.0-0 \
libxtst6 \
libxss1 \
libgconf-2-4 \
libnss3 \
libasound2 && \
mkdir -p /home/orca && \
cd /home/orca && \
wget https://github.com/plotly/orca/releases/download/v1.2.1/orca-1.2.1-x86_64.AppImage && \
chmod +x orca-1.2.1-x86_64.AppImage && \
./orca-1.2.1-x86_64.AppImage --appimage-extract && \
printf '#!/bin/bash \nxvfb-run --auto-servernum --server-args "-screen 0 640x480x24" /home/orca/squashfs-root/app/orca "$@"' > /usr/bin/orca && \
chmod +x /usr/bin/orca
# enabling xvfb
import plotly.io as pio
pio.orca.config.use_xvfb = True
pio.orca.config.save()
Please see the example yaml file at script_configs/pred.yml. There's a tumor-only
example in script_configs/infer_tu_only/pred.yml.
# kidney-tumor
python /content/kits19-cnn/scripts/predict.py --yml_path="/content/kits19-cnn/script_configs/pred.yml"
# tumor only
python /content/kits19-cnn/scripts/predict.py --yml_path="/content/kits19-cnn/script_configs/infer_tu_only/pred.yml"
Please see the example yaml file at script_configs/eval.yml. There's a tumor-only
example in script_configs/infer_tu_only/eval.yml.
# kidney-tumor
python /content/kits19-cnn/scripts/evaluate.py --yml_path="/content/kits19-cnn/script_configs/eval.yml"
# tumor only
python /content/kits19-cnn/scripts/evaluate.py --yml_path="/content/kits19-cnn/script_configs/infer_tu_only/eval.yml"
Currently, only on the preprocess-test-set branch.