This is the repository of our solution to the 2020 edition of the BraTS challenge. Our paper is available at arXiv.
This repository implements Pipeline A training and inference only. Feel free to use it as a starter for following challenge editions!
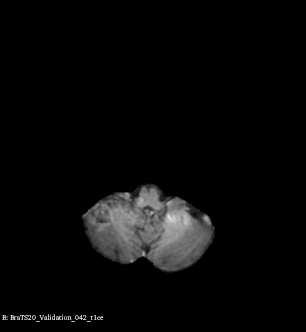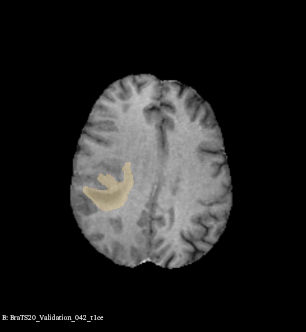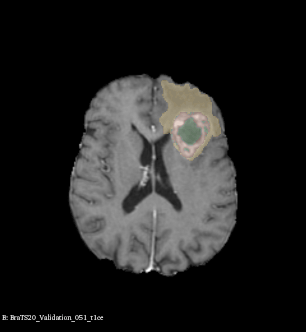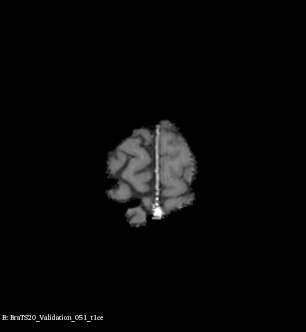
Example of brain tumor segmentations generated by our solution
in your favorite virtual environment:
git clone https://github.com/lessw2020/Ranger-Deep-Learning-Optimizer
cd Ranger-Deep-Learning-Optimizer
pip install -e .
cd open_brats2020
pip install -r requirements.txtFirst change your data source folder by modifying values in src/config.py
BRATS_TRAIN_FOLDERS = "your-Path_to/brats2020/MICCAI_BraTS_2020_Data_Training"
BRATS_VAL_FOLDER = "your-Path_to/brats2020/MICCAI_BraTS_2020_Data_Valdation"
BRATS_TEST_FOLDER = "your-Path_to/brats2020/MICCAI_BraTS_2020_Data_Testing"If you prefer not to hardcode this value, you can set them as variable environments.
Then, start training:
python -m src.train --devices 0 --width 48 --arch EquiUnet
For more details on the available option:
python -m src.train -h
Note that this code use an nn.Module to do data augmentation: any batch size above 1 could lead to errors.
After training, you will have a runs folder created containing a directory for each run you have done.
For each run, a yaml file with the option used for the runs, and
a segs folder containing the generated .nii.gz segmentations for the validation fold used.
- src
- runs
- 20201127_34335135__fold_etc
202020201127_34335135__fold_etc.ymal
- segs
model.txt # the printed model
model_best.pth.tar # model weights
patients_indiv_perf.csv # a log of training patient segmentation performance
events.out.. # Tensorboard log
The yaml file is required to perform inference on the validation and train set
The script to perform inference is... inference.py !!
python -m src.inference -h
usage: inference.py [-h] [--config PATH [PATH ...]] --devices DEVICES
[--on {val,train,test}] [--tta] [--seed SEED]
Brats validation and testing dataset inference
optional arguments:
-h, --help show this help message and exit
--config PATH [PATH ...]
path(s) to the trained models config yaml you want to
use
--devices DEVICES Set the CUDA_VISIBLE_DEVICES env var from this string
--on {val,train,test}
--tta
--seed SEED
This script can take multiple models (specify multiple yaml config files), even when trained with different image normalization techniques (minmax or zscore); and will automatically merge their prediction (by averaging).