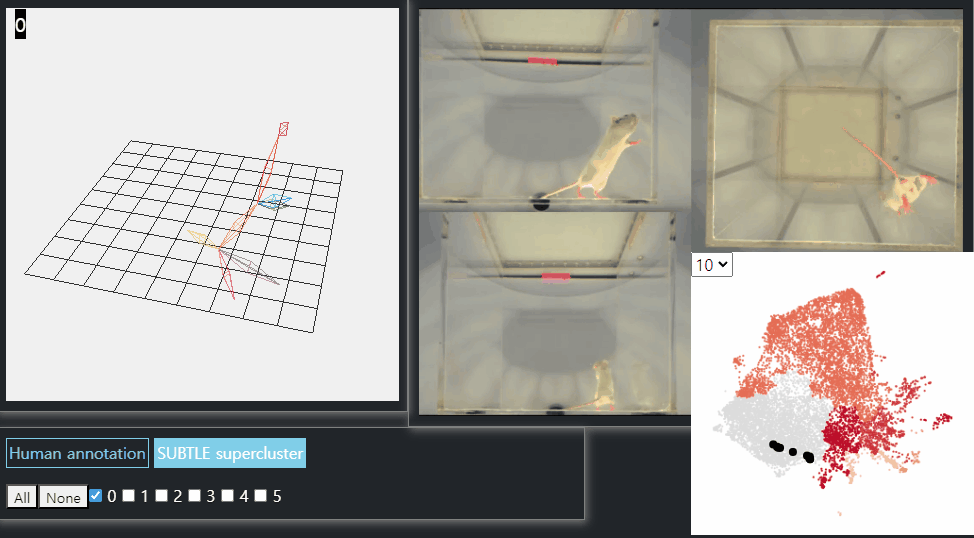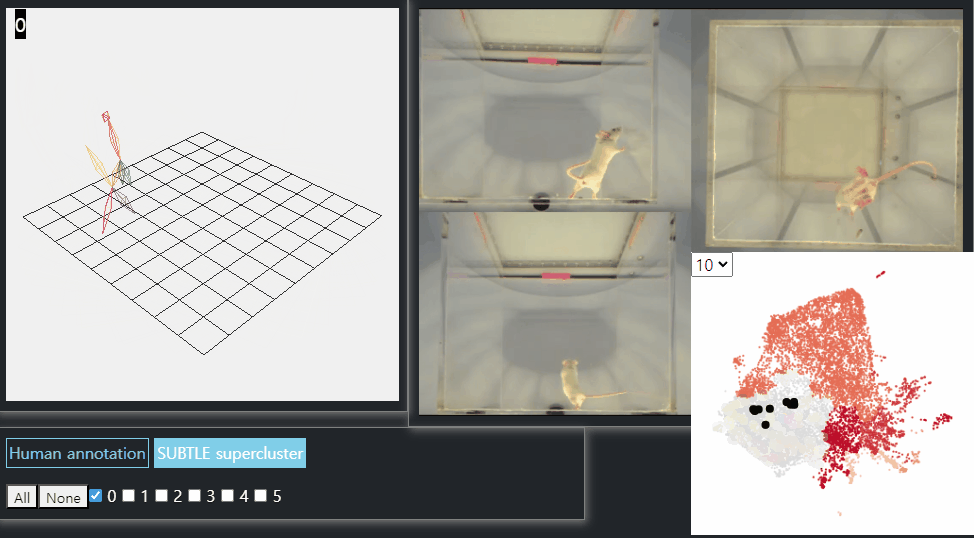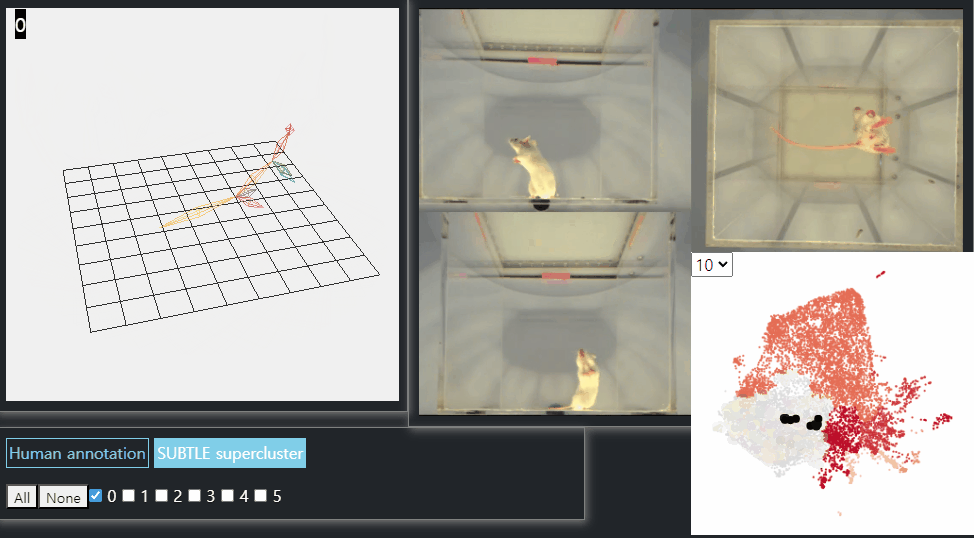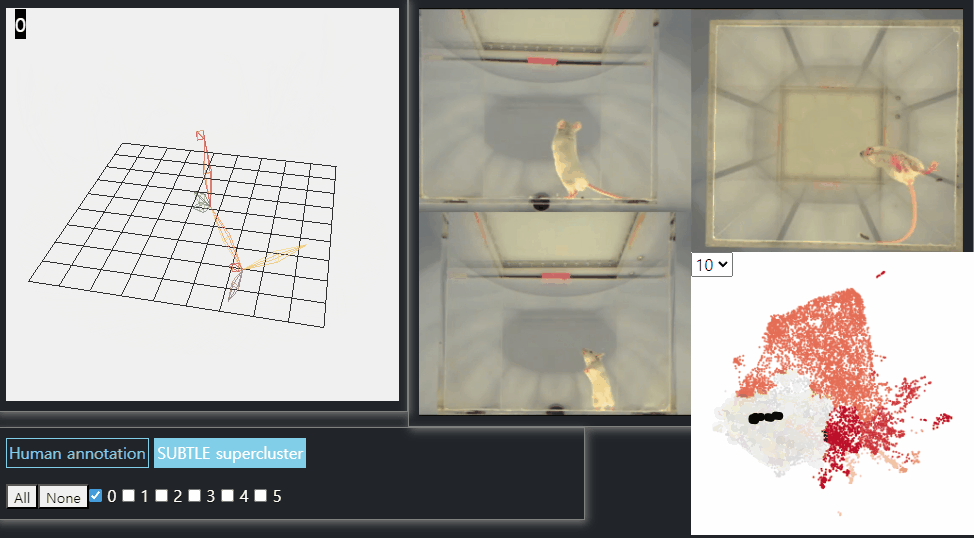
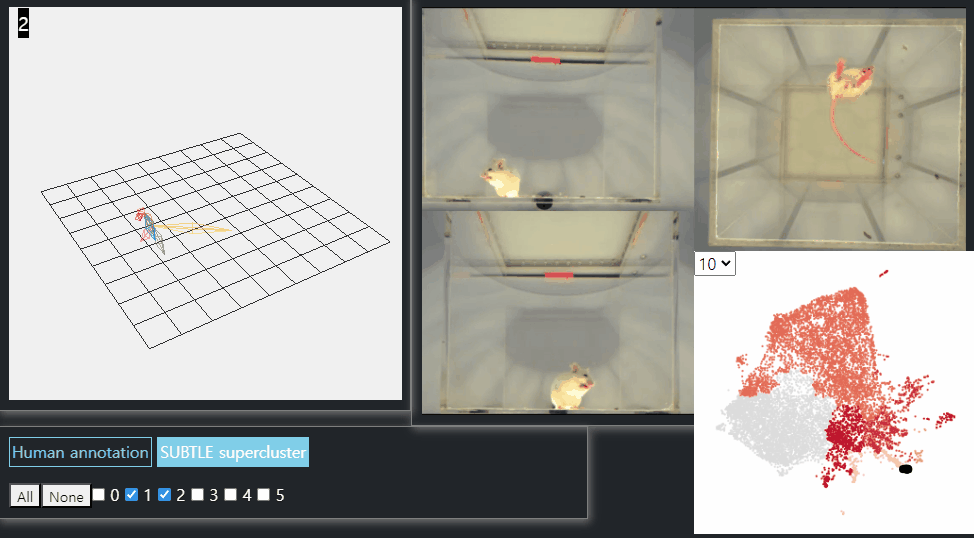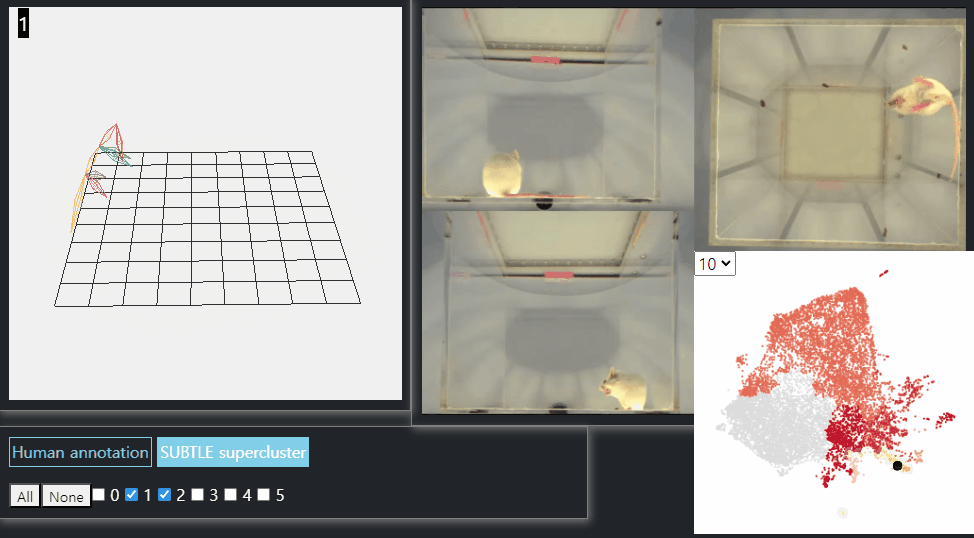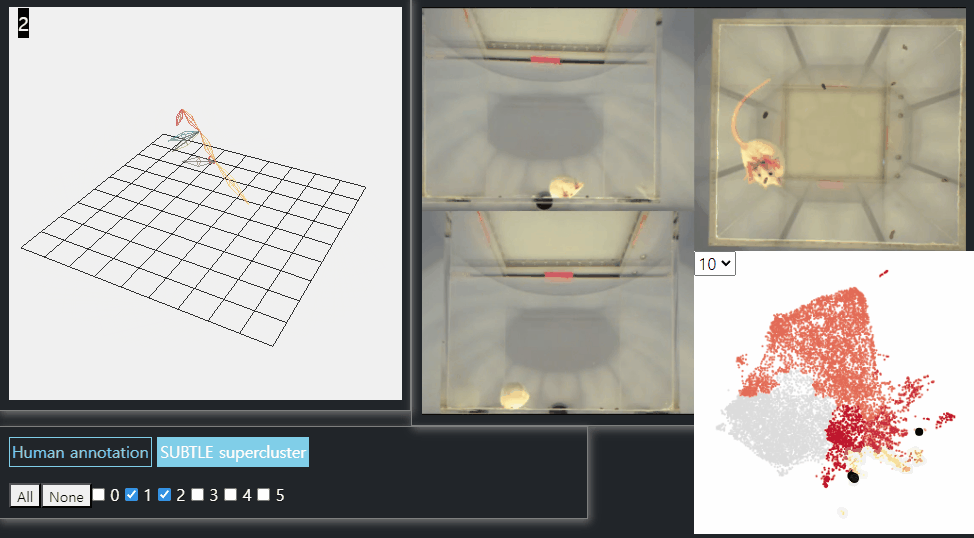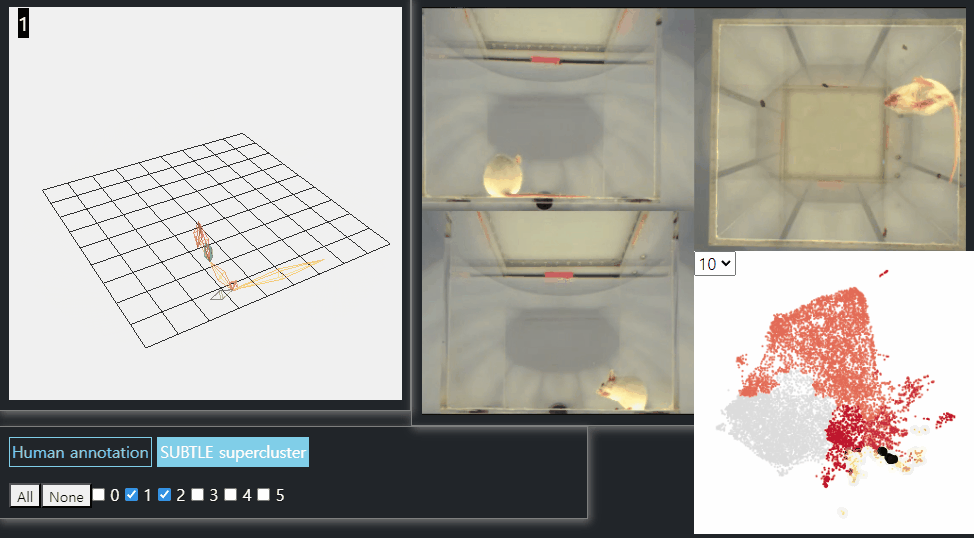
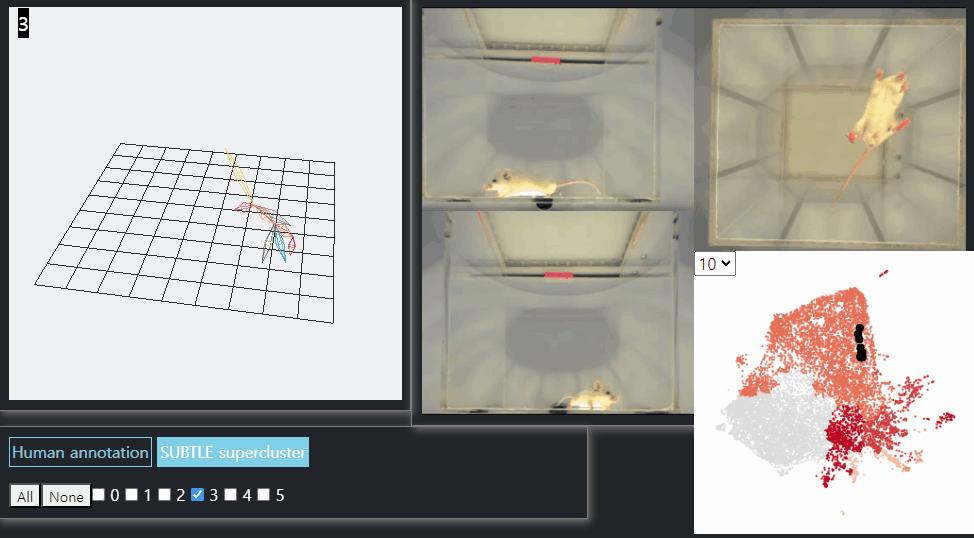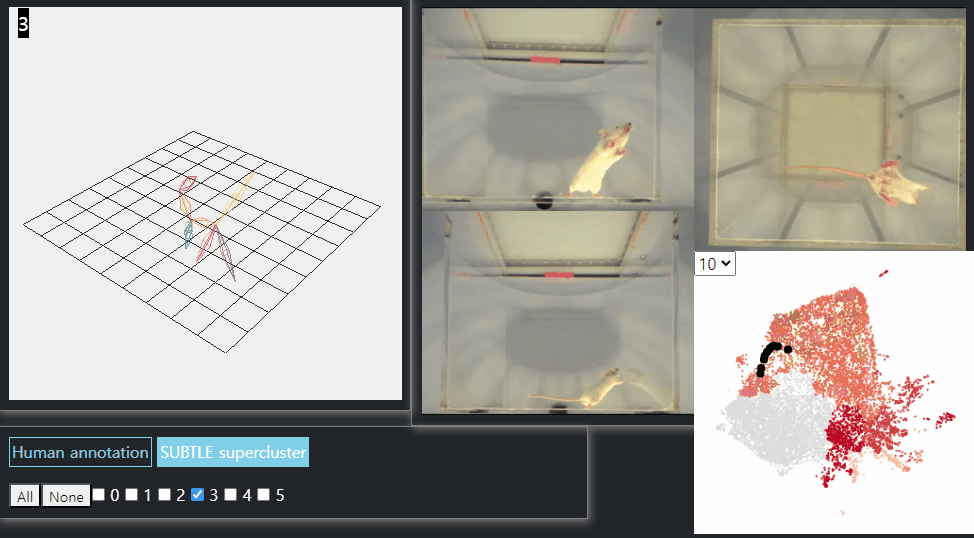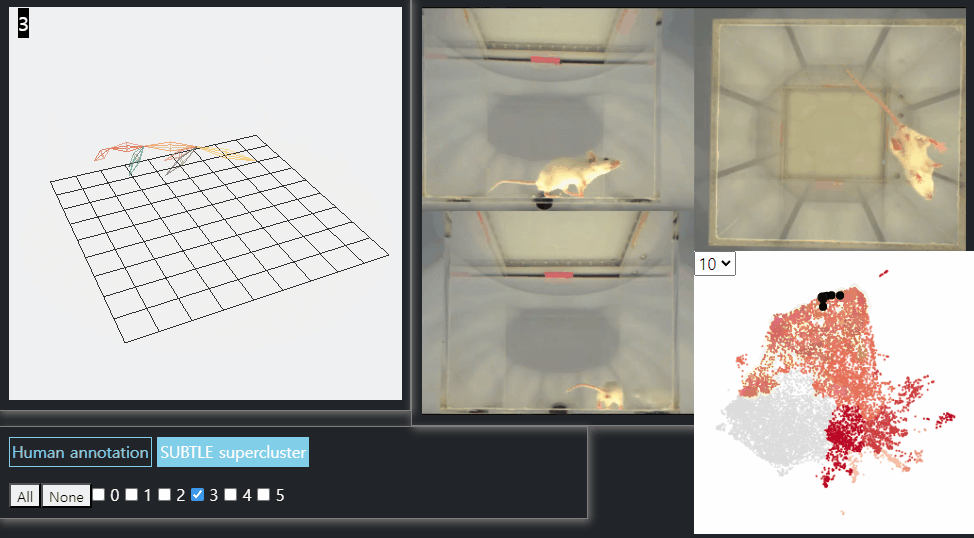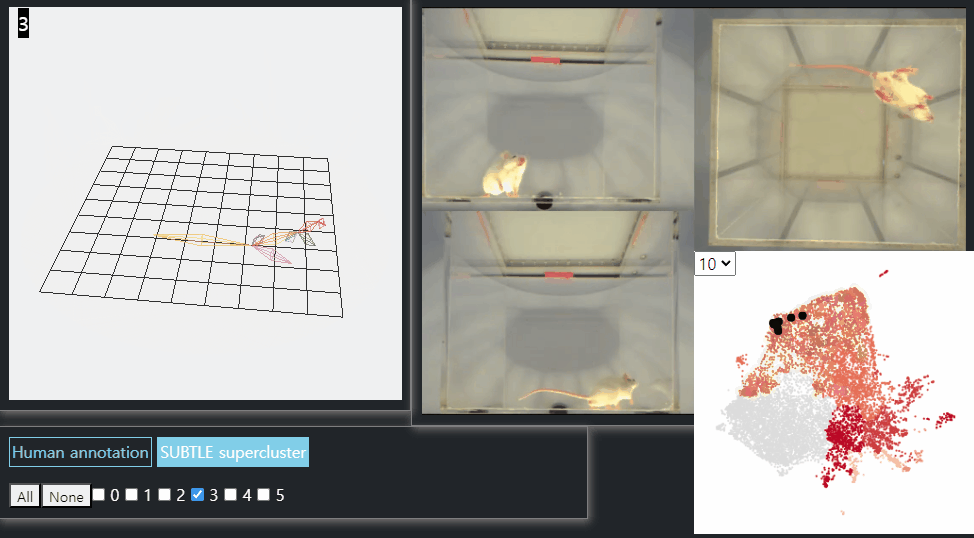
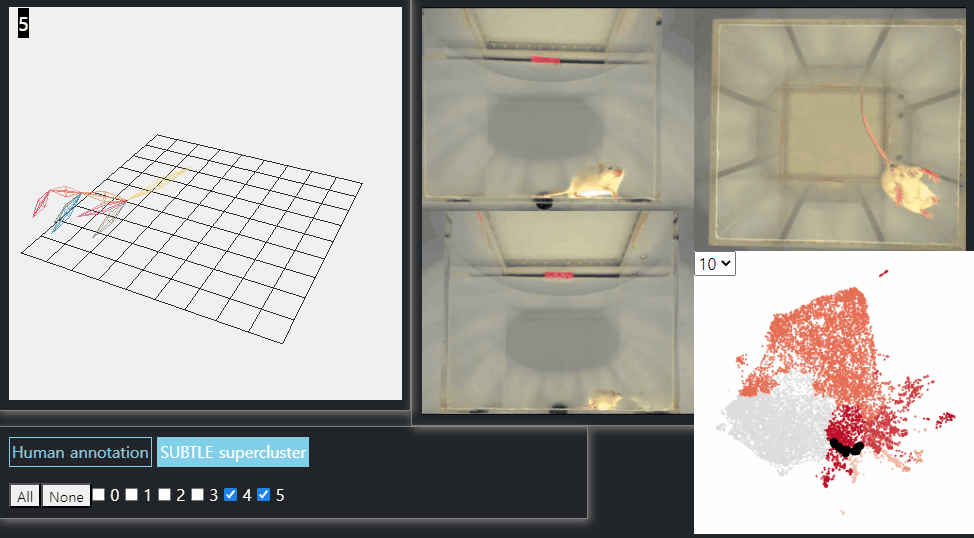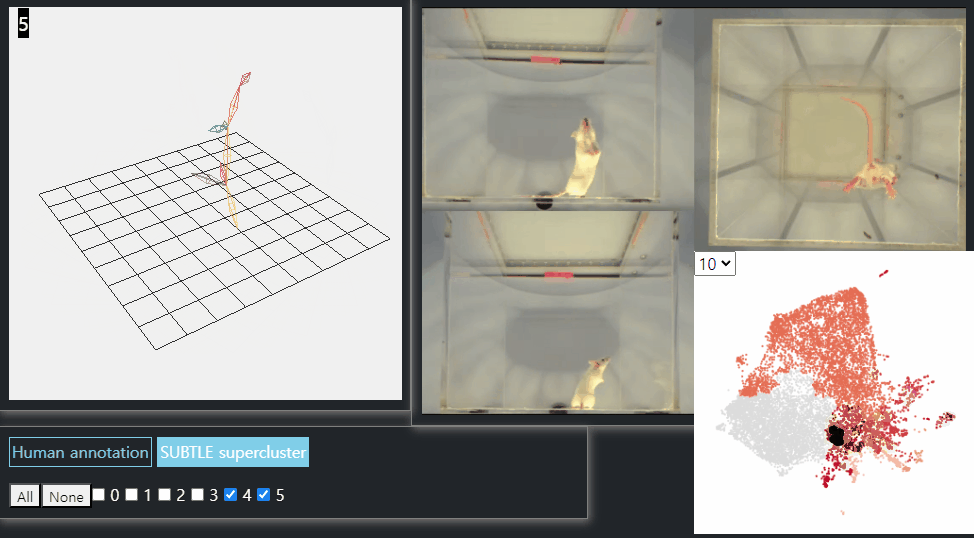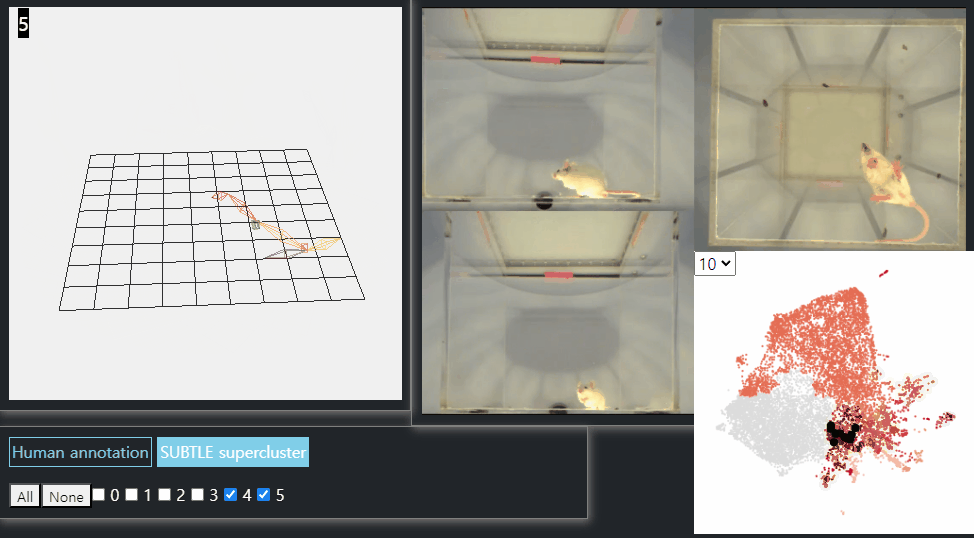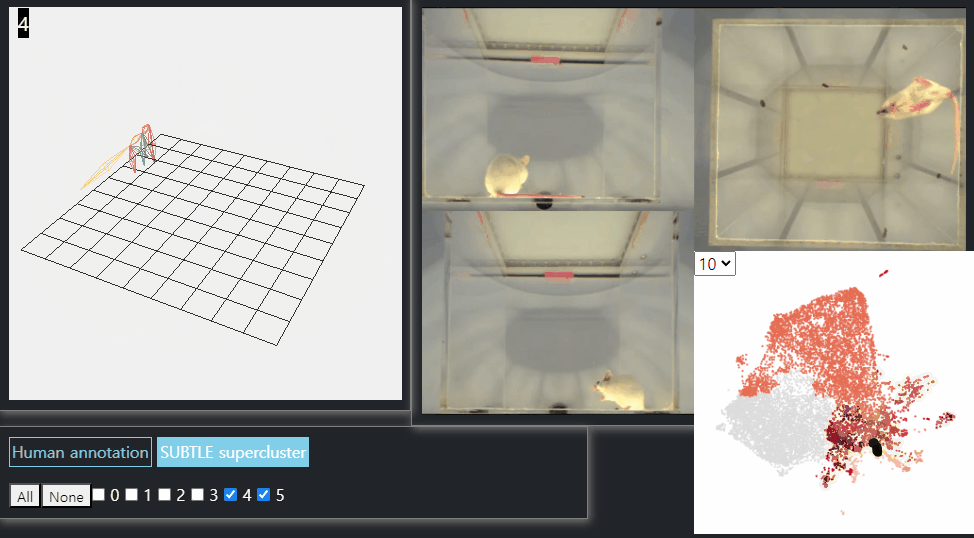
SUBTLE framework
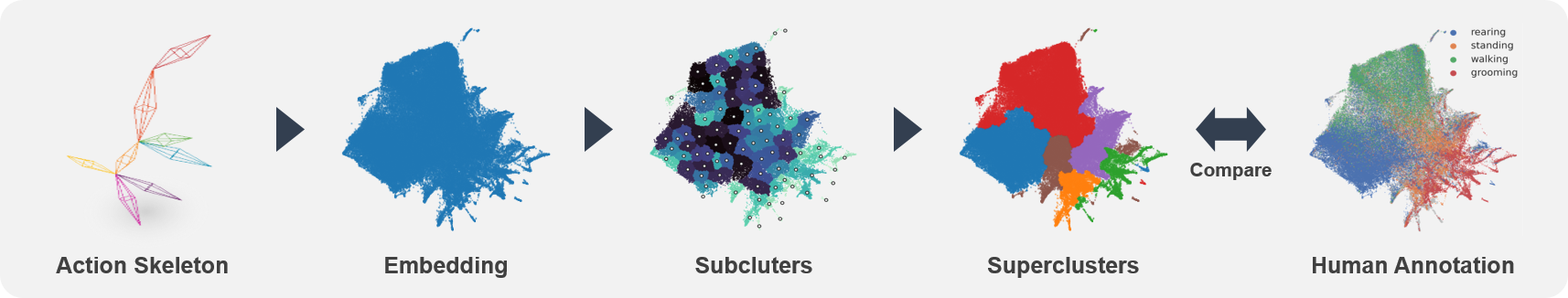 SUBTLE automates the tasks of both identifying behavioral repertoires such as walking, grooming, standing, and rearing from freely moving mice. Our framework utilizes Spectrogram-UMAP-based Temporal Link Embedding (SUBTLE) which effectively reflects both temporal and kinematic representation in the behavioral embedding space. From this embeding space, we create subclusters as behavioral states, which serve as building blocks for identifying superclusters as behavioral repertoires. For more details, see our paper.
SUBTLE automates the tasks of both identifying behavioral repertoires such as walking, grooming, standing, and rearing from freely moving mice. Our framework utilizes Spectrogram-UMAP-based Temporal Link Embedding (SUBTLE) which effectively reflects both temporal and kinematic representation in the behavioral embedding space. From this embeding space, we create subclusters as behavioral states, which serve as building blocks for identifying superclusters as behavioral repertoires. For more details, see our paper.
Visualization (Website)
Check out our website for more information and a GUI web page for SUBTLE framework.
Rearing-like behavior repertoires (supercluster 0)
Grooming-like behavior repertoires (supercluster 1, 2)
Walking-like behavior repertoires (supercluster 3)
Standing-like behavior repertoires (supercluster 4, 5)
Installation
pip install -U git+https://github.com/jeakwon/subtle.git
or
pip install -U https://github.com/jeakwon/subtle/archive/refs/heads/main.zip
Datasets
Access our (3d action skeleton datasets)[https://github.com/jeakwon/subtle/tree/main/dataset].
- List of files : 19 action skeleton recordings 10 human annotations
- Frame length : ~12,000 frames
- Sampling rate : 20 fps
- Recording time : ~10 minutes
- Recording system : AVATAR system paper@biorxiv, poster@cv4animals, contact@hompage
- Annotated labels : walking, rearing, standing, grooming, na (not assigned)
Quick Demo
You can try simple demonstration in colab
Prepare dataset
import subtle
import pandas as pd
# Dataset for training (5 young 5 adult mice)
y5a5 = [
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/adult_6112.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/adult_6115.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/adult_6116.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/adult_6127.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/adult_7678.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/young_7100.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/young_7678.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/young_8294.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/young_8296.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y5a5/coords/young_8301.csv',
]
# Dataset for mapping (3 young 6 adult mice)
y3a6 = [
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8294.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8296.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8301.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8765.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8767.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/adult_8789.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/young_8765.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/young_8767.csv',
'https://raw.githubusercontent.com/jeakwon/subtle/main/dataset/y3a6/coords/young_8789.csv',
]
training_dataset = []
training_nameset = []
for csv in y5a5:
X = pd.read_csv(csv, header=None).values
X = subtle.avatar_preprocess(X) # subtract coordinates with global mean of (x, y, z)
training_dataset.append(X)
training_name = csv.split('/')[-1].replace('.csv', '')
training_nameset.append(training_name)
mapping_dataset = []
mapping_nameset = []
for csv in y3a6:
X = pd.read_csv(csv, header=None).values
X = subtle.avatar_preprocess(X) # subtract coordinates with global mean of (x, y, z)
mapping_dataset.append(X)
mapping_name = csv.split('/')[-1].replace('.csv', '')
mapping_nameset.append(mapping_name)
Training and Mapping
mapper = subtle.Mapper(fs=20) # fs, sampling frequency
training_outputs = mapper.fit(training_dataset)
mapping_outputs = mapper.run(mapping_dataset)Save and Load trained model
mapper.save('trained_model.pkl')
mapper = subtle.load('trained_model.pkl')Save output result into csv files
for name, output in zip(training_nameset, training_outputs):
# export embeddings
df = pd.DataFrame(output.Z)
df.to_csv(name+'_embeddings.csv', header=None, index=None)
# export subclusters
df = pd.DataFrame(output.y)
df.to_csv(name+'_subclusters.csv', header=None, index=None)
# export superclusters
df = pd.DataFrame(output.Y)
df.to_csv(name+'_superclusters.csv', header=None, index=None)
# export transition probabilities
df = pd.DataFrame(output.TP)
df.to_csv(name+'transition_probabilities.csv', header=None, index=None)
# export retention rate
df = pd.DataFrame(output.R)
df.to_csv(name+'retention_rate.csv', header=None, index=None)Visualize trained result
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].scatter(mapper.Z[:, 0], mapper.Z[:, 1], s=1, c=mapper.y) # subclusters
ax[1].scatter(mapper.Z[:, 0], mapper.Z[:, 1], s=1, c=mapper.Y[:, -1]) # superclusters