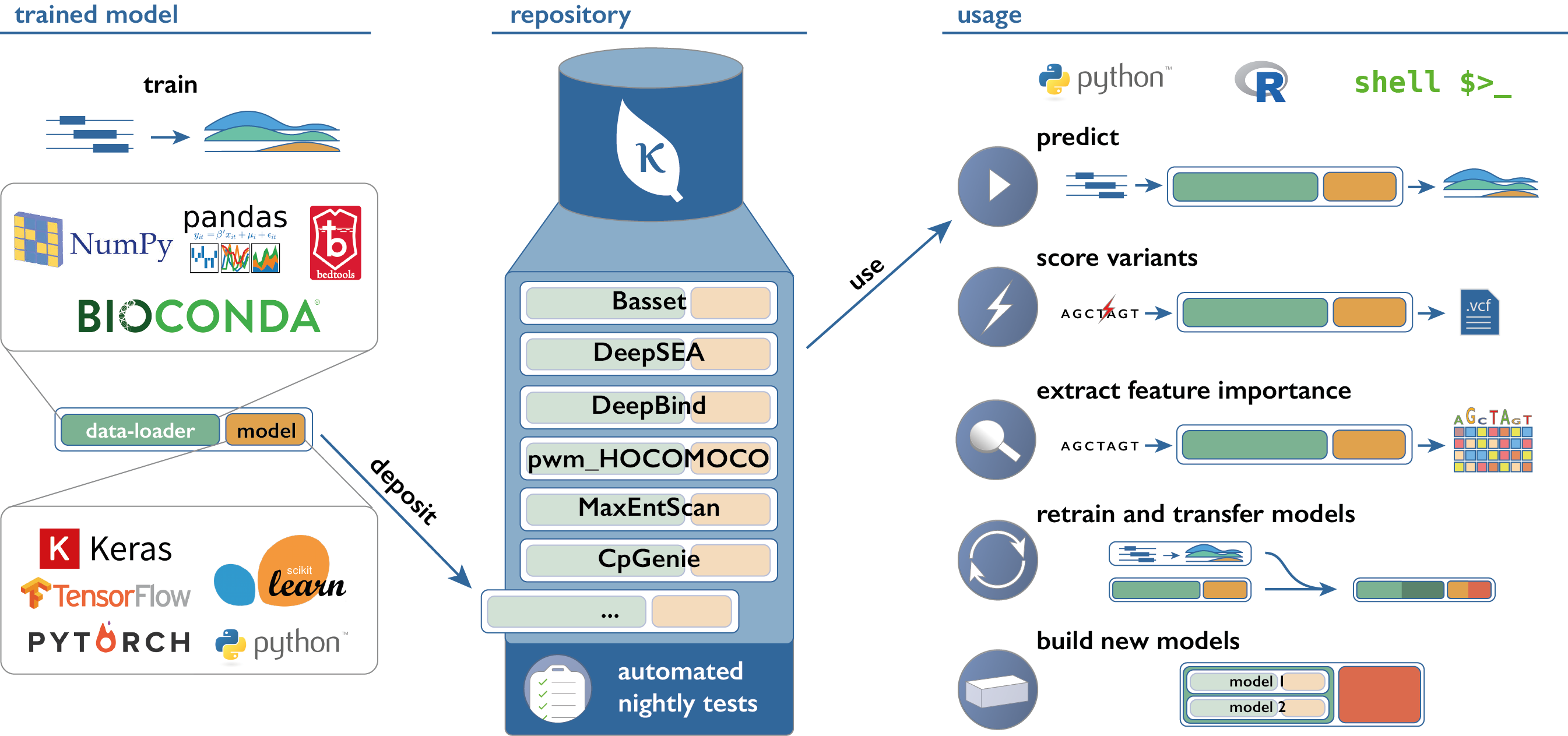
This repository implements a python package and a command-line interface (CLI) to access and use models from Kipoi-compatible model zoo's.
- kipoi.org - Main website
- kipoi.org/docs - Documentation
- github.com/kipoi/models - Model zoo for genomics maintained by the Kipoi team
Kipoi requires conda to manage model dependencies. Make sure you have either anaconda (download page) or miniconda (download page) installed. If you are using OSX, see Installing python on OSX.
For downloading models, Kipoi uses git and Git Large File Storage (LFS). See how to install git here. To install git-lfs on Ubuntu, run:
curl -s https://packagecloud.io/install/repositories/github/git-lfs/script.deb.sh | sudo bash
sudo apt-get install -y git git-lfs
git-lfs installAlternatively, you can install git-lfs through conda:
conda install -c conda-forge git-lfs && git lfs installNext, install Kipoi using pip:
pip install kipoiIf you wish to develop kipoi, run instead:
conda install pytorch-cpu
pip install -e '.[develop]'This will install some additional packages like pytest. You can test the package by running py.test.
If you wish to run tests in parallel, run py.test -n 6.
List available models
import kipoi
kipoi.list_models()Hint: For an overview over the available models also check the model overview on our website, where you can see example commands for how to use the models on the CLI, python and R.
Load the model from model source or local directory
# Load the model from github.com/kipoi/models/rbp
model = kipoi.get_model("rbp_eclip/UPF1", source="kipoi") # source="kipoi" is the default
# Load the model from a local directory
model = kipoi.get_model("~/mymodels/rbp", source="dir")
# Note: Custom model sources are defined in ~/.kipoi/config.yaml
# Load the model via github permalink for a particular commit:
model = kipoi.get_model("https://github.com/kipoi/models/tree/7d3ea7800184de414aac16811deba6c8eefef2b6/pwm_HOCOMOCO/human/CTCF", source='github-permalink')Main model attributes and methods
# See the information about the author:
model.info
# Access the default dataloader
model.default_dataloader
# Access the Keras model
model.model
# Predict on batch - implemented by all the models regardless of the framework
# (i.e. works with sklearn, Keras, tensorflow, ...)
model.predict_on_batch(x)
# Get predictions for the raw files
# Kipoi runs: raw files -[dataloader]-> numpy arrays -[model]-> predictions
model.pipeline.predict({"dataloader_arg1": "inputs.csv"})Load the dataloader
Dl = kipoi.get_dataloader_factory("rbp_eclip/UPF1") # returns a class that needs to be instantiated
dl = Dl(dataloader_arg1="inputs.csv") # Create/instantiate an objectDataloader attributes and methods
# batch_iter - common to all dataloaders
# Returns an iterator generating batches of model-ready numpy.arrays
it = dl.batch_iter(batch_size=32)
out = next(it) # {"inputs": np.array, (optional) "targets": np.arrays.., "metadata": np.arrays...}
# To get predictions, run
model.predict_on_batch(out['inputs'])
# load the whole dataset into memory
dl.load_all()Re-train the model
# re-train example for Keras
dl = Dl(dataloader_arg1="inputs.csv", targets_file="mytargets.csv")
it_train = dl.batch_train_iter(batch_size=32)
# batch_train_iter is a convenience wrapper of batch_iter
# yielding (inputs, targets) tuples indefinitely
model.model.fit_generator(it_train, steps_per_epoch=len(dl)//32, epochs=10)For more information see: nbs/python-api.ipynb and docs/using getting started
$ kipoi
usage: kipoi <command> [-h] ...
# Kipoi model-zoo command line tool. Available sub-commands:
# - using models:
ls List all the available models
predict Run the model prediction
pull Download the directory associated with the model
preproc Run the dataloader and save the results to an hdf5 array
postproc Tools for model postprocessing like variant effect prediction
env Tools for managing Kipoi conda environments
# - contribuing models:
init Initialize a new Kipoi model
test Runs a set of unit-tests for the model
test-source Runs a set of unit-tests for many/all models in a source
Explore the CLI usage by running kipoi <command> -h. Also, see docs/using/getting started cli for more information.
You can add your own (private) model sources. See docs/using/03_Model_sources/.
See docs/contributing getting started and docs/tutorials/contributing/models for more information.
Functionality to predict the effect of SNVs is available in the API as well as in the command line interface. The input is a VCF which can then be annotated with effect predictions and returned in the process. For more details on the requirements for the models and dataloaders please check docs/using/02_Variant_effect_prediction
Documentation can be found here: kipoi.org/docs