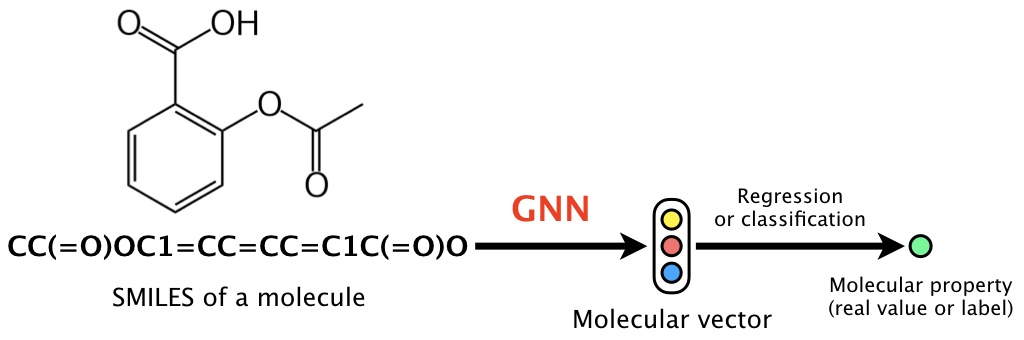
This is the code of a graph neural network (GNN) for molecules, which is based on learning representations of r-radius subgraphs (or fingerprints) in molecules. This GNN is proposed in our paper "Compound-protein Interaction Prediction with End-to-end Learning of Neural Networks for Graphs and Sequences (Bioinformatics, 2018)," which aims to predict compound-protein interactions for drug discovery. Using the proposed GNN, in this page we provide an implementation of the model for predicting various molecular properties such as drug efficacy and photovoltaic efficiency.
- This code is easy to use. After setting the environment (e.g., PyTorch), preprocessing data and learning a model can be done by only two commands (see "Usage").
- If you prepare a molecular property dataset with the same format as provided in the dataset directory, you can learn our GNN with your dataset by the two commands (see "Training of our GNN using your molecular property dataset").
The basic idea of a GNN can be described as follows:
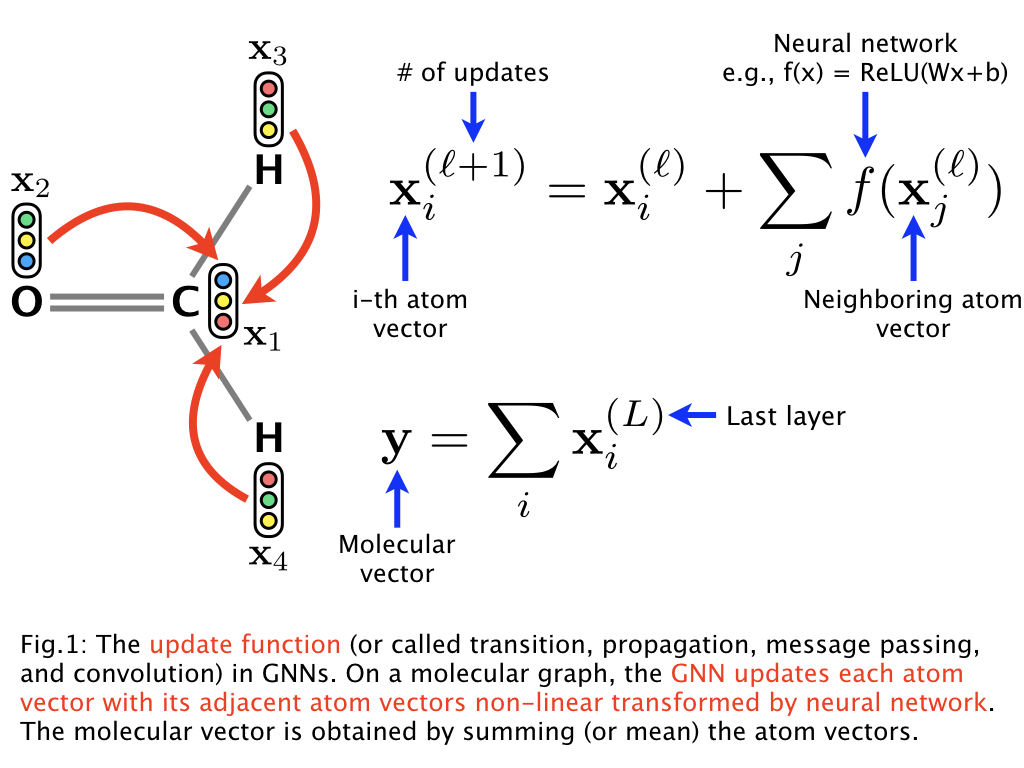
The GNN (1) updates the randomly initialized atom vectors considering the graph structure of a molecule, (2) obtains the molecular vector, and then (3) learns the neural network parameters including the atom vectors via backpropagation to predict a molecular property. That is, this is the end-to-end learning that does not require input features or descriptors used in chemoinformatics.
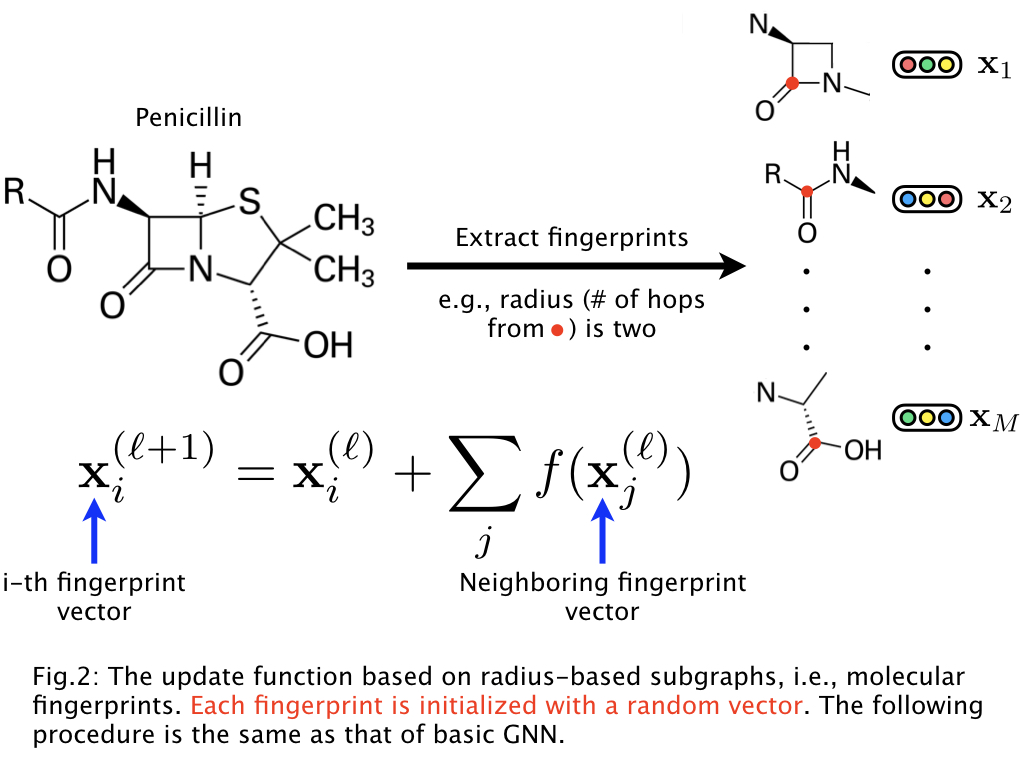
In drug compounds, for example, each atom, chemical bond, and their connections in a molecular graph are not so important. More important in drug compounds is to consider relatively large fragments in a molecular graph, e.g., β-lactam in penicillin. Such fragments are referred to as r-radius subgraphs or molecular fingerprints. Based on this observation, our GNN leverages molecular fingerprints and the model can be described as follows:
Thus, instead of using atom vectors, we (1) extract the fingerprints from a molecular graph and initialize them using random vectors, (2) obtain the molecular vector by GNN, and then (3) learn the representations. This GNN allows us to learn the representations of molecular fingerprints.
- PyTorch
- scikit-learn
- RDKit
We provide two major scripts:
-
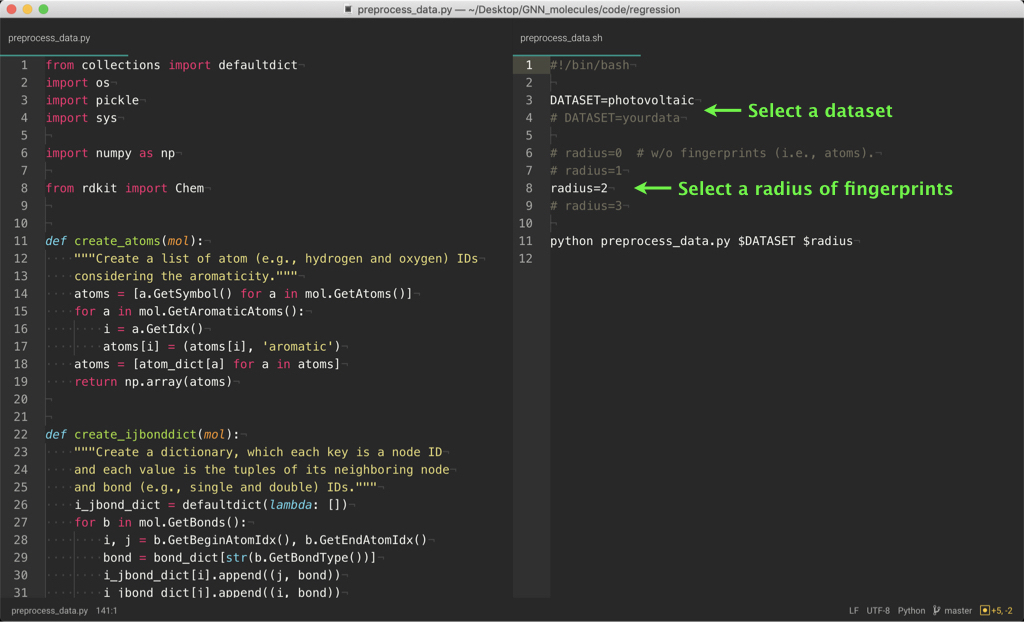
code/regression or classification/preprocess_data.py creates the input tensor data of molecules for processing with PyTorch from the original data (see dataset/regression or classification/original/data.txt).
-
code/regression or classification/run_training.py trains our GNN using the above preprocessed data to predict a molecular property.
Create the tensor data of molecules and their properties with the following command:
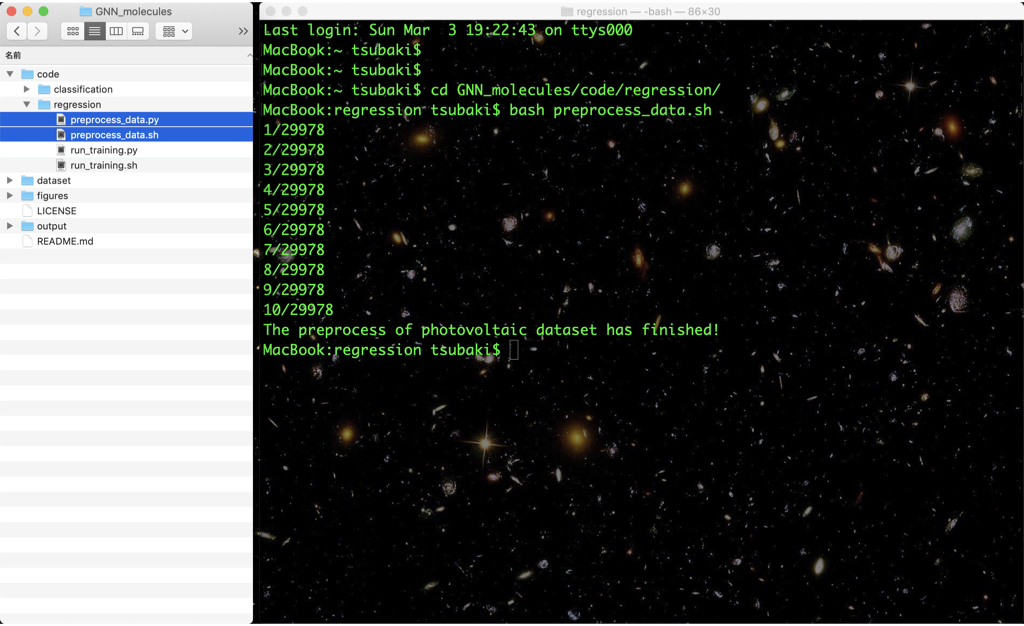
cd code/regression (or cd code/classification)
bash preprocess_data.sh
The preprocessed data are saved in the dataset/input directory.
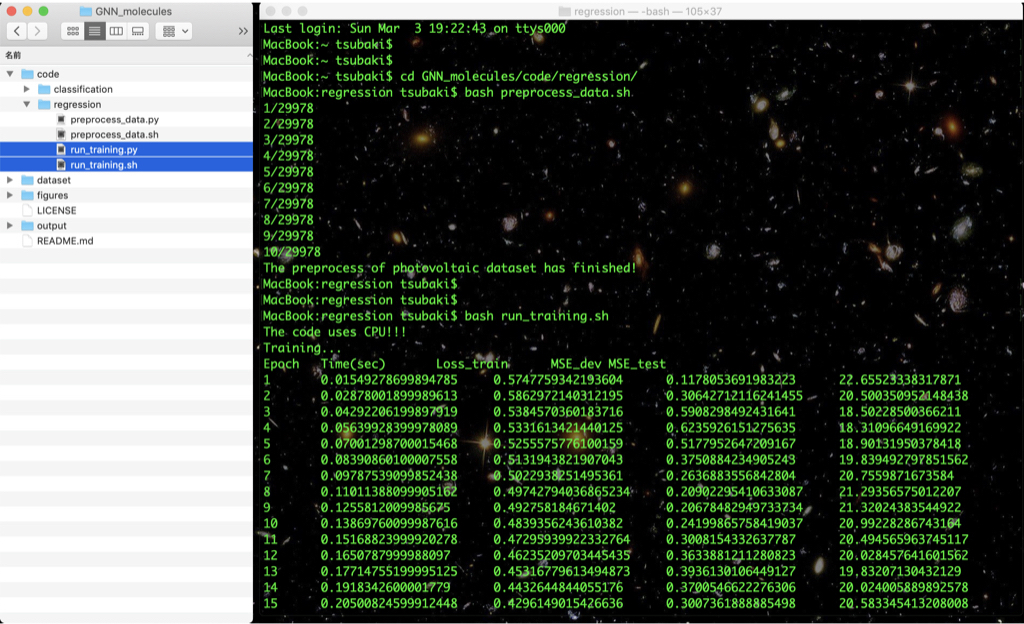
Using the preprocessed data, train our GNN with the following command:
bash run_training.sh
The training and test results and the GNN model are saved in the output directory (after training, see output/result and output/model).
You can change the hyperparameters in preprocess_data.sh and run_training.sh as described in the above figures. Try to learn various models!
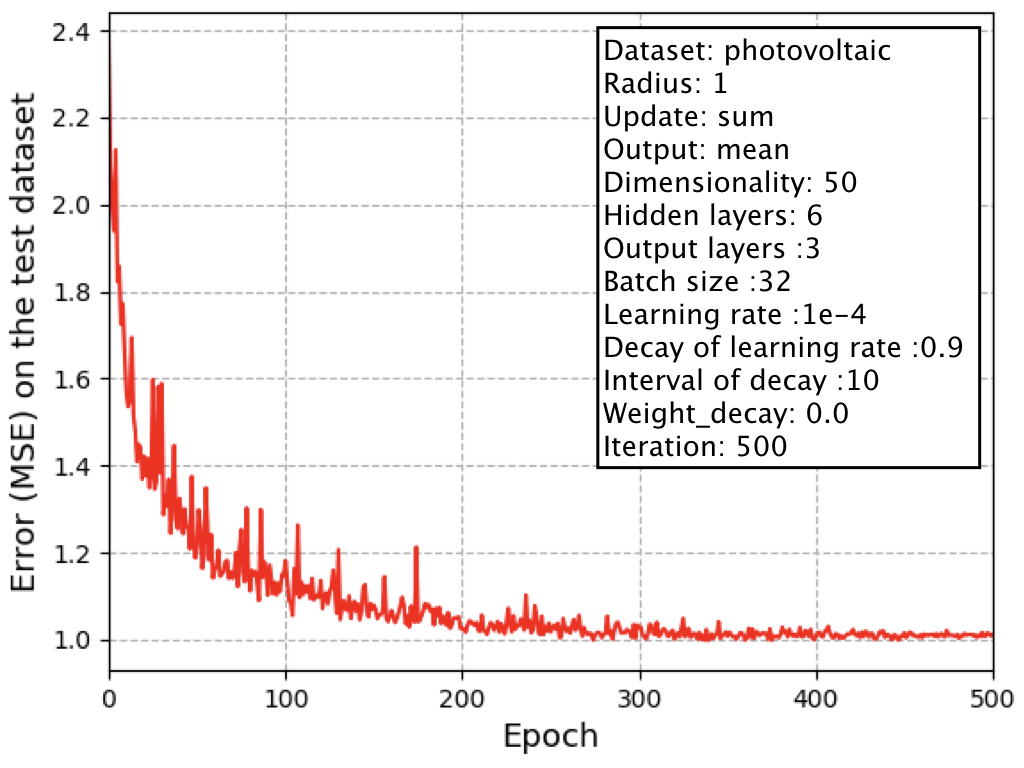
On the photovoltaic efficiency dataset in the directory of dataset/regression, the learning curve (x-axis is epoch and y-axis is error) is as follows:
This result can be reproduce by the above two commands (1) and (2).
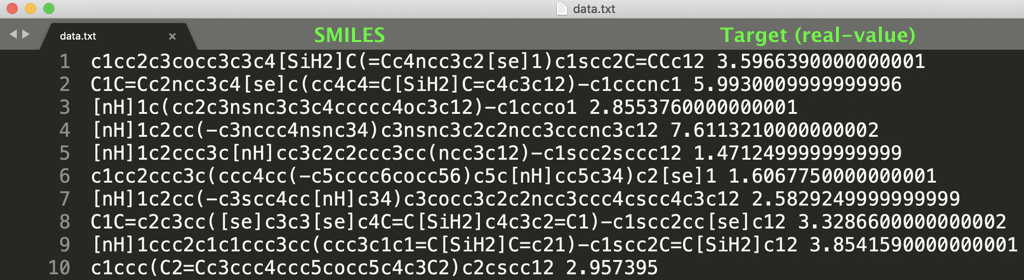
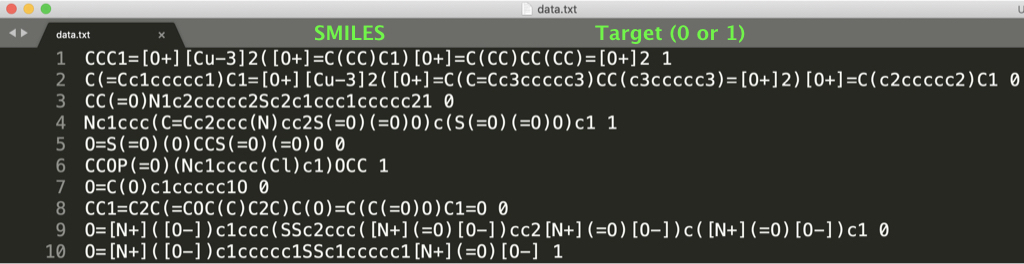
In this repository, we provide two datasets of regression (see dataset/regression/photovoltaic/original/data.txt) and classification (see dataset/classification/HIV/original/data.txt) as follows:
If you prepare a dataset with the same format as "data.txt" in a new directory (e.g., dataset/yourdata/original), you can train our GNN using your dataset by the above two commands (1) and (2).
- Preprocess data contains "." in the SMILES format (i.e., a molecule contains multi-graphs).
- Provide some pre-trained models and the demo scripts.
@article{tsubaki2018compound,
title={Compound-protein Interaction Prediction with End-to-end Learning of Neural Networks for Graphs and Sequences},
author={Tsubaki, Masashi and Tomii, Kentaro and Sese, Jun},
journal={Bioinformatics},
year={2018}
}