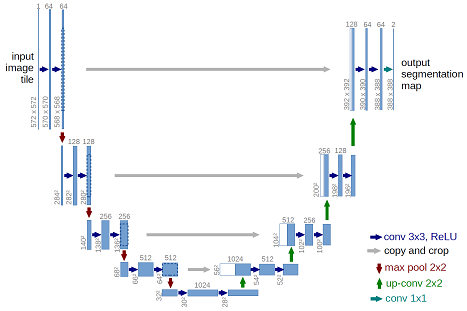
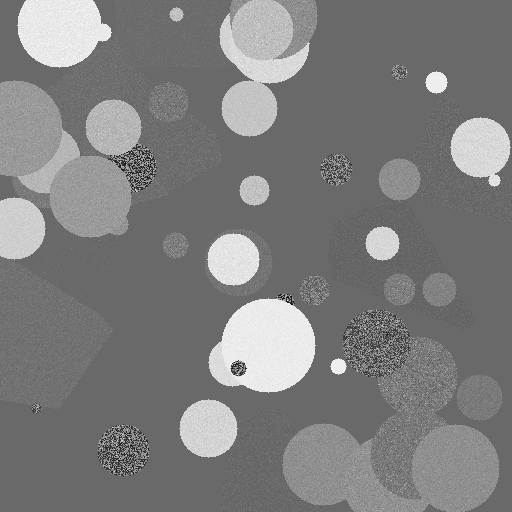
A rigorous method for automated soft tissue seg-mentation using planar kilovoltage (kV) imaging, a photoncounting detector (PCD), and a convolutional neural network ispresented. The goal of the project was to determine the optimumnumber of energy bins in a PCD for soft tissue segmentation.Planar kV X-ray images of solid water (SW) phantoms withvarying depth of cartilage were generated with a cone-beamanalytical method and parallel-beam Monte Carlo simulations.Simulations were preformed using 1 to 5 PCD energy bins withequal photon fluence distribution. Simulated image signal tonoise ratio (SNR) was varied between 10 to 250 measured aftertransmission through 4cm of SW. Algorithms using non-linearas well as linear regression were used to predict the amount ofcartilage for every pixel of the phantom. These algorithms wereevaluated based on the mean squared error (MSE) between theirprediction and the ground truth. The best algorithm was usedto decompose randomly generated SW and cartilage images withan SNR of 100. These randomly generated images trained a U-Net convolutional neural network to segment the cartilage in theimage. The results indicated the smallest MSE occurred for non-linear regression with 4 energy bins over all SNR. The trainedU-Net was able to correctly segment all regions of cartilage forthe smallest amount of cartilage used (4 mm) and segmented theregion with >99% categorical accuracy by pixel.
Code for the Monte Carlo simulation, matlab analysis, image generation, and unet CNN can be found here!
First clone the repository into the directory of your choice,
Using mac or linux:
git clone https://github.com/jerichooconnell/unet.git
This tutorial depends on the following libraries:
- Tensorflow
- Keras >= 1.0
Also, this code should be compatible with Python versions 2.7-3.5.
The -gpu should change to -cpu if you are using a cpu Assuming jupyter is already installed:
virtualenv tensorflow
source tensorflow/bin/activate
pip install tensorflow-gpu keras==2.1.6 scikit-image
If you are using a gpu you should also install cuda and cudNN
If you are using a cluster with slurm workload manager, and interactive nodes (bit of a long shot) your workflow could look like this: These commands will set up a jupyter environment
module purge
module load python/3.5
virtualenv tensorflow2
source tensorflow2/bin/activate
module load cuda cudNN
pip install tensorflow-gpu keras==2.1.6 scikit-image
pip install jupyter
echo -e '#!/bin/bash\nunset XDG_RUNTIME_DIR\njupyter notebook --ip $(hostname -f) --no-browser' > $VIRTUAL_ENV/bin/notebook.sh
This command will start a server on an interactive node, you would need your own account though
salloc --time=1:0:0 --ntasks=1 --cpus-per-task=6 --gres=gpu:1 --mem=32000M --account=def-bazalova srun $VIRTUAL_ENV/bin/notebook.sh'''
A message with information about where the server is running will appear with a node number to put in <###> Then on a local terminal create a tunnel to
ssh -f jerichoo@cedar.computecanada.ca -L 8890:cdr<###>.int.cedar.computecanada.ca:8888 -N
The jupyter server should be forwarded to your browser at
https://localhost/8888
Install TOPAS which may or may not be free
- TOPAS - MC simulation framework
Excecute the REDLEN_many_runs.sh script from one of the run directories
Install matlab which is definetly not be free
There are two directories MC_3.0 which contains the analytical analysis which allows arbitrary SNRs but is limited in energy bins and DES_MC which is the monte carlo code which allows arbitrary binning but is limited in SNR
Both methods have a Main which calls the functions. There are three binary variables which control whether or not the linear method is used and/or the nonlinear method is used. Initialize loads new data but expects a certain directory structure.
Again install matlab which is definetly not be free
Further, install the matlab API for python, sorry that I use this but I really wanted a matlab function.
The notebook image_generation.ipynb will take you through the image generation process.
The architecture was inspired by U-Net: Convolutional Networks for Biomedical Image Segmentation.
This implimentation is a combination of zhixuhao's unet implimentation with a generator borrowed heavily from jakeret's tf_unet implementation.
The data is randomly generated CT images using a 4 energy PCD
The data for training contains 30 512*512 images, which are far not enough to feed a deep learning neural network. I use a module called ImageDataGenerator in keras.preprocessing.image to do data augmentation.
See dataPrepare.ipynb and data.py for detail.
This deep neural network is implemented with Keras functional API, which makes it extremely easy to experiment with different interesting architectures.
Output from the network is a 512*512 which represents mask that should be learned. Sigmoid activation function makes sure that mask pixels are in [0, 1] range.
The model is trained for 5 epochs.
After 5 epochs, calculated accuracy is about 0.99.
Loss function for the training is basically just a binary crossentropy.
Use the trained model to do segmentation on test images, the result is statisfactory.
Keras is a minimalist, highly modular neural networks library, written in Python and capable of running on top of either TensorFlow or Theano. It was developed with a focus on enabling fast experimentation. Being able to go from idea to result with the least possible delay is key to doing good research.
Use Keras if you need a deep learning library that:
allows for easy and fast prototyping (through total modularity, minimalism, and extensibility). supports both convolutional networks and recurrent networks, as well as combinations of the two. supports arbitrary connectivity schemes (including multi-input and multi-output training). runs seamlessly on CPU and GPU. Read the documentation Keras.io
Keras is compatible with: Python 2.7-3.5.