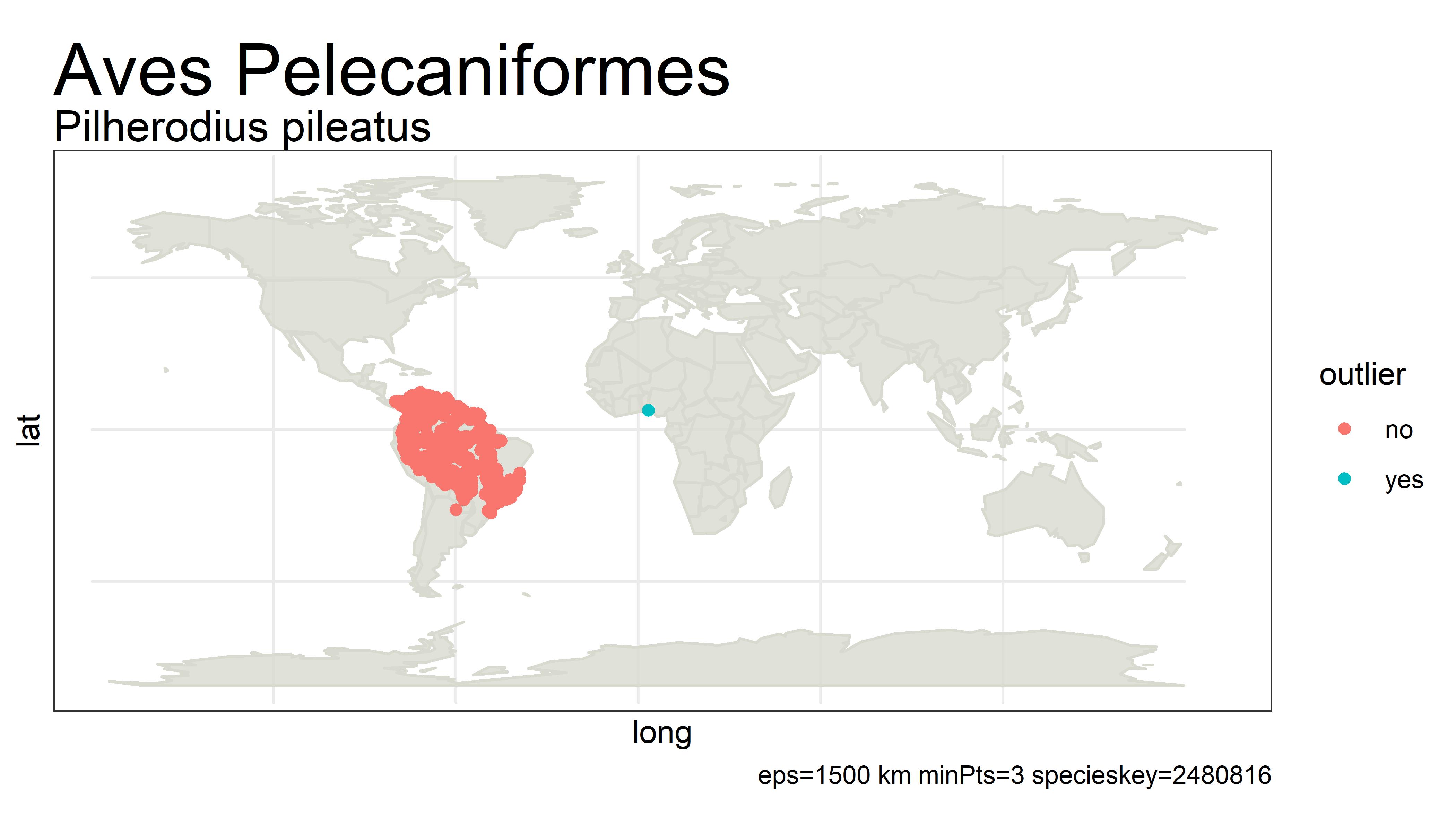
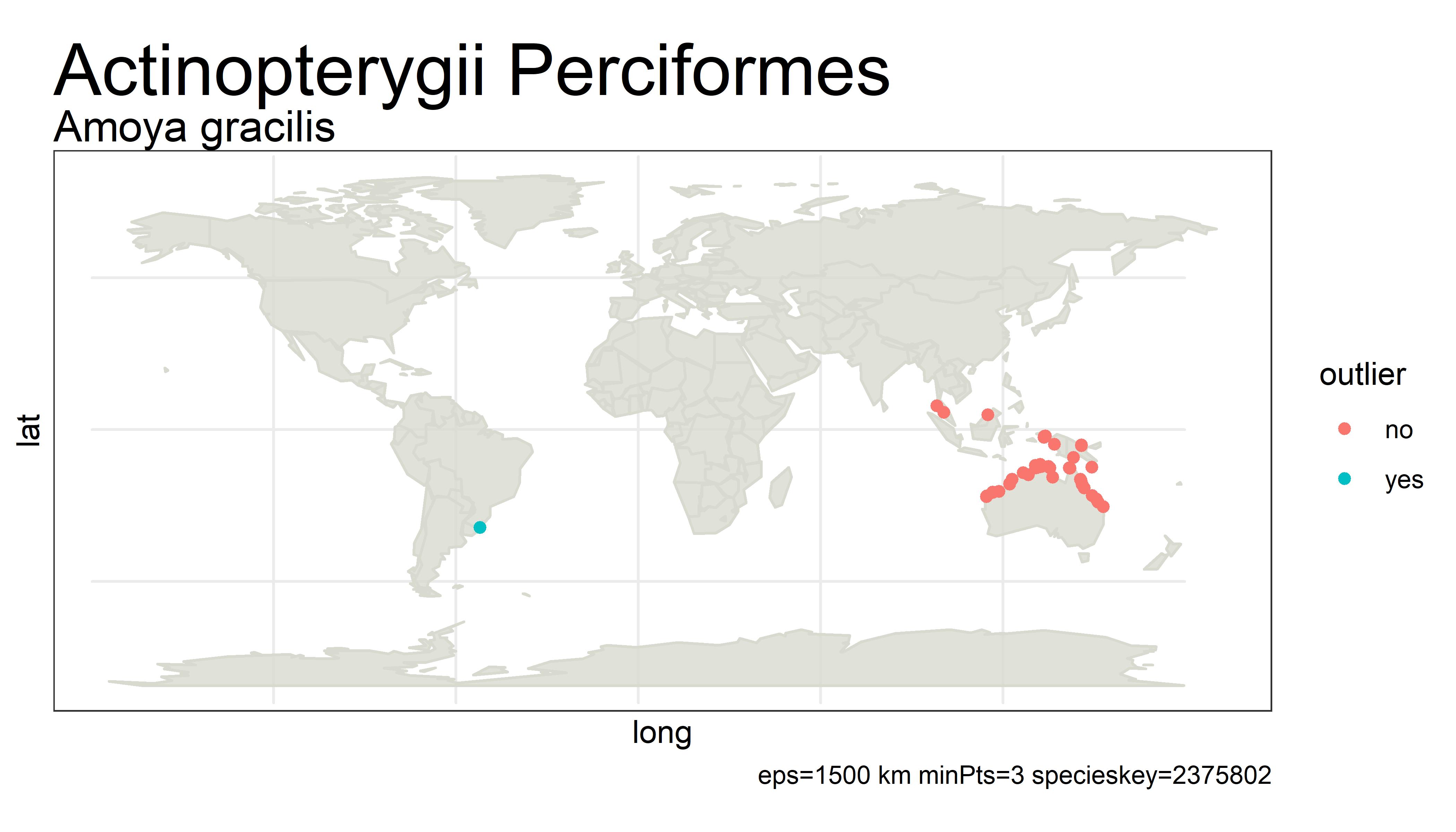
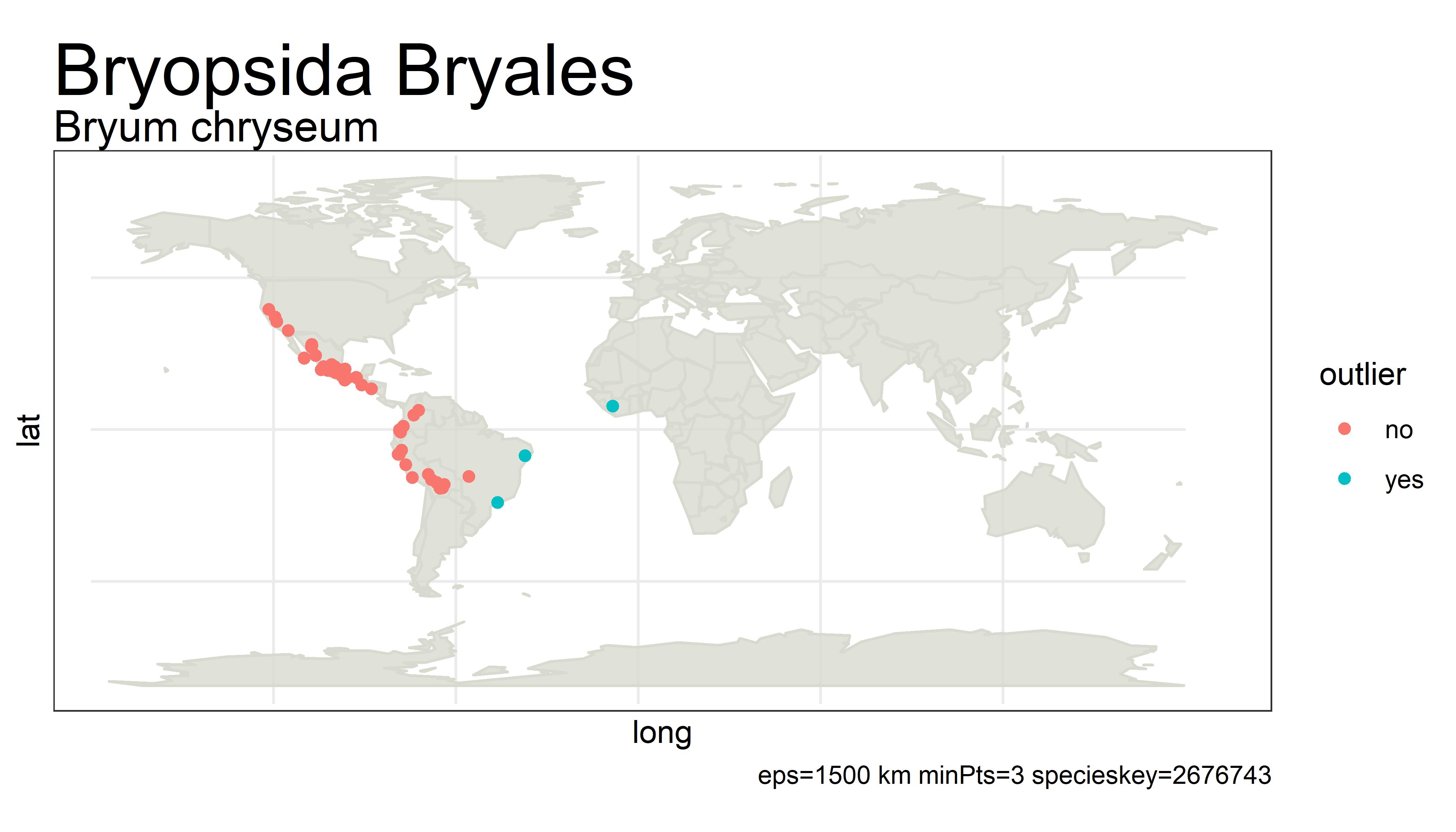
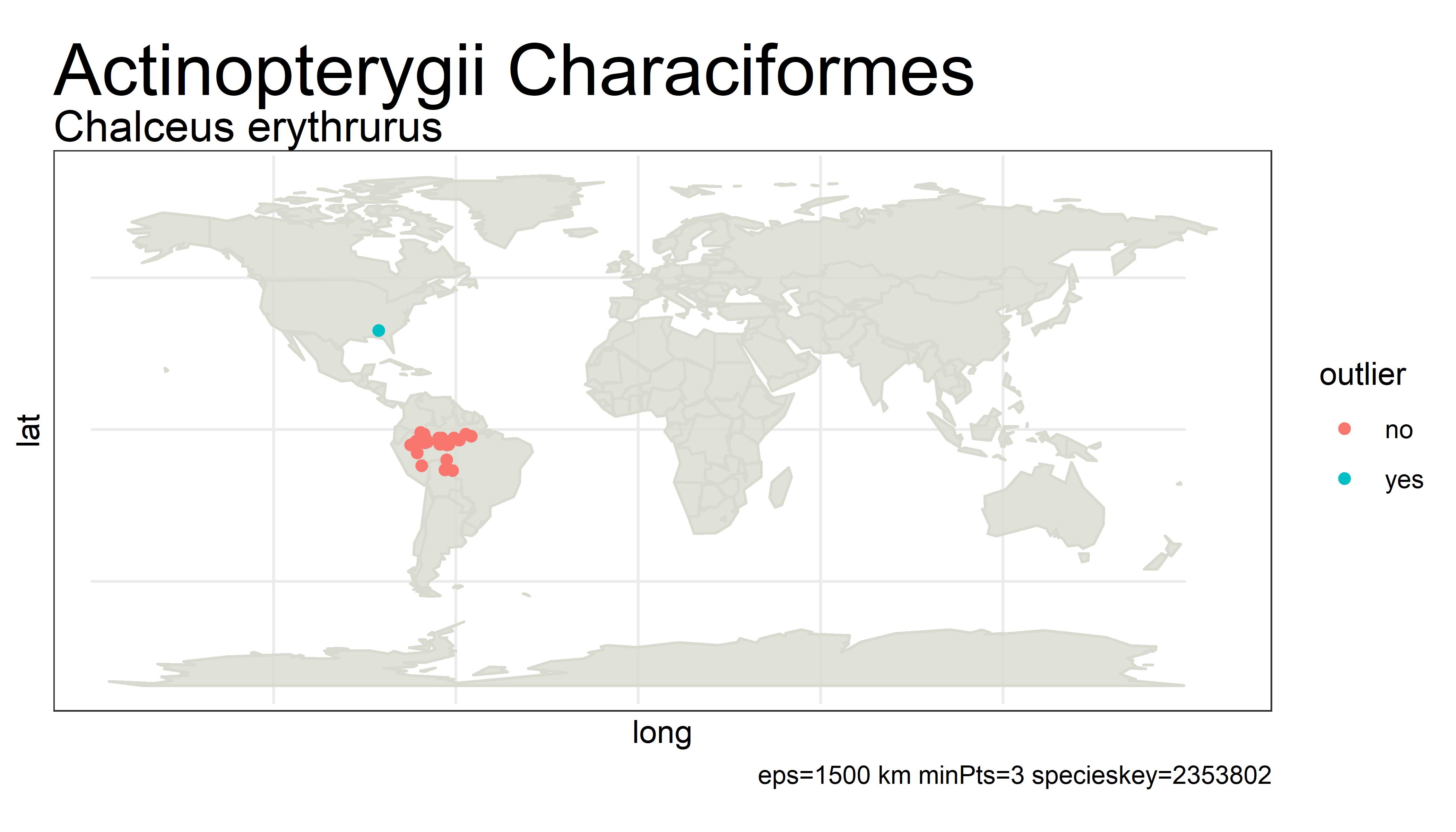
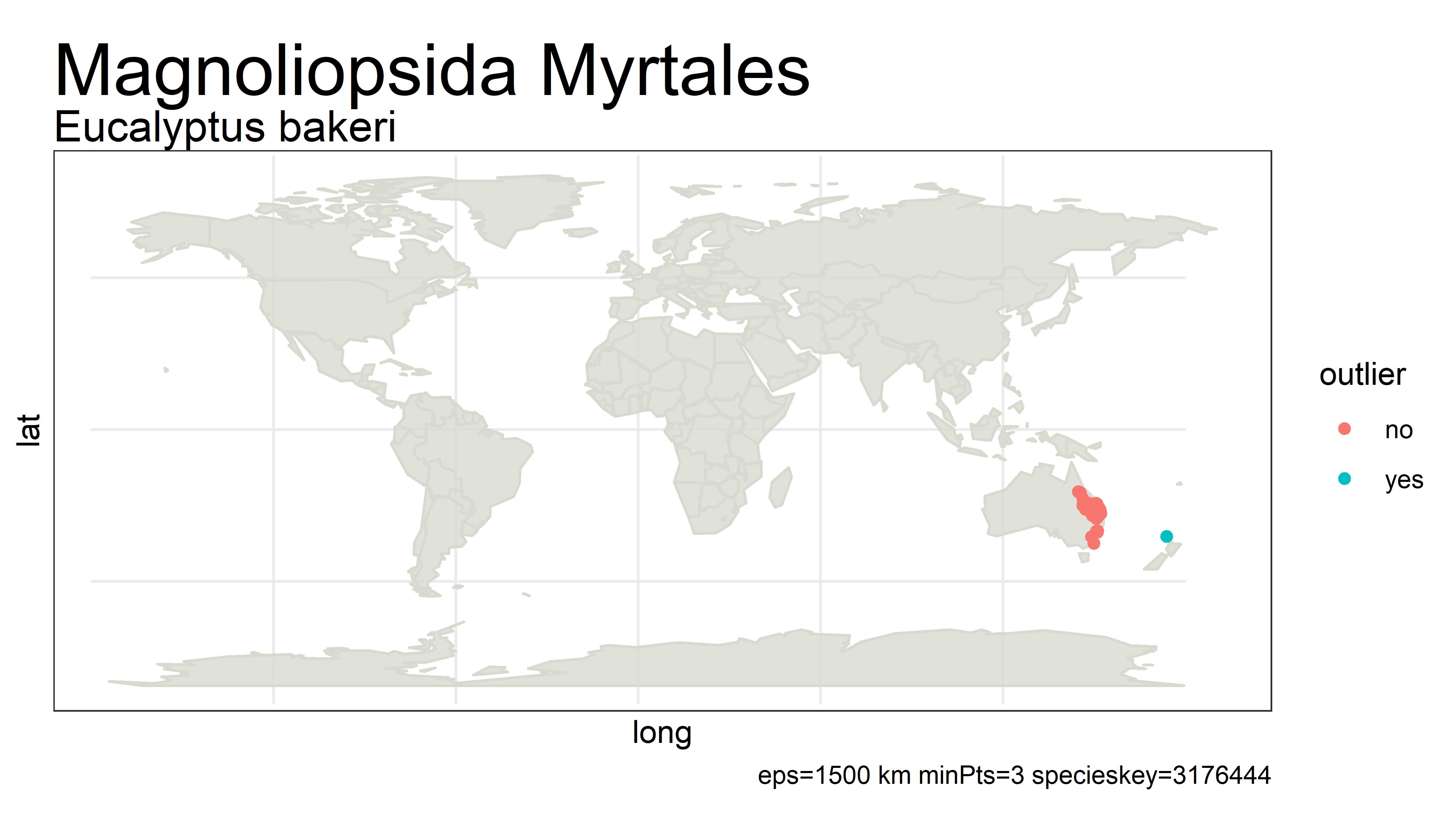
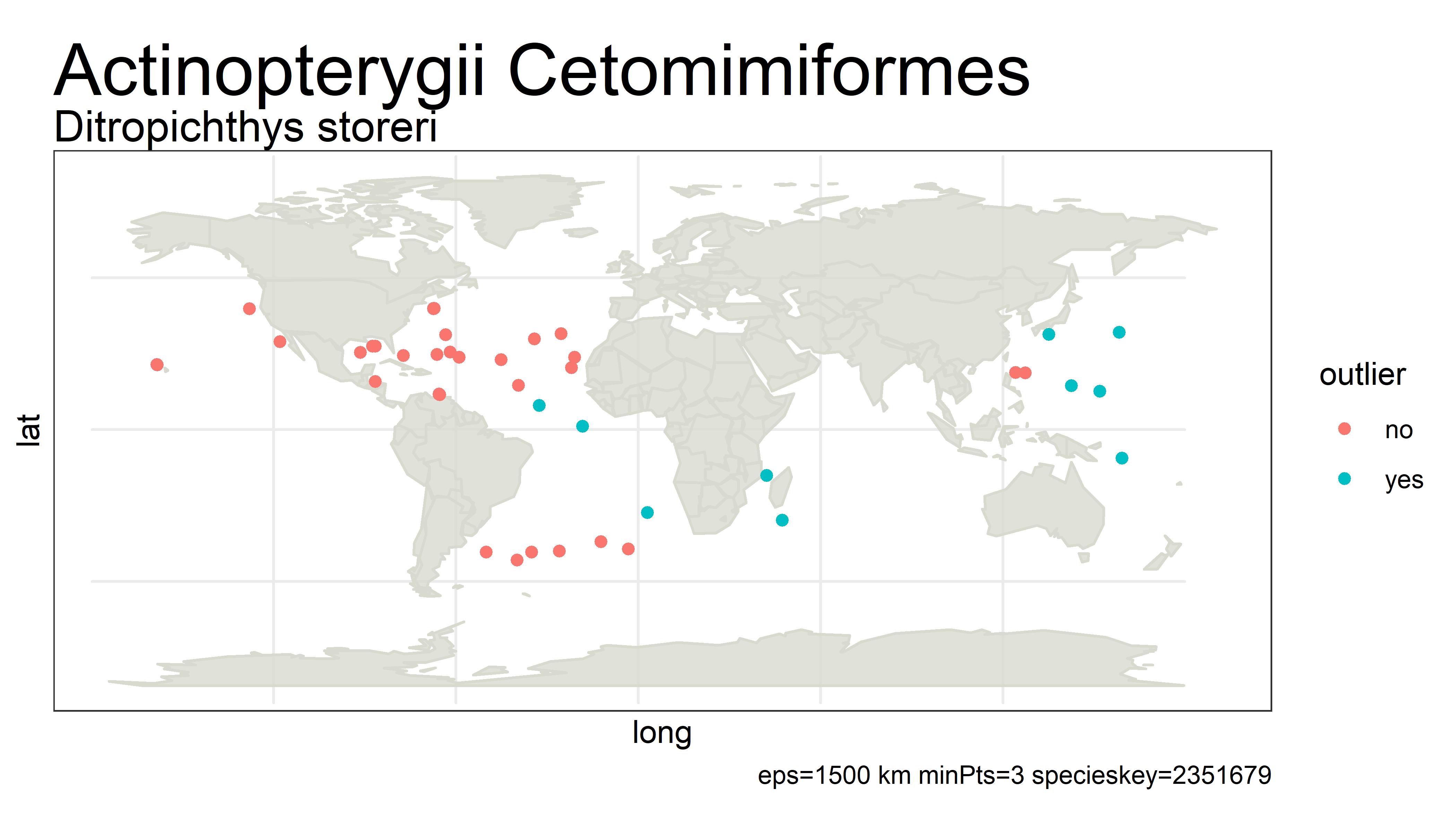
This project detects outliers in GBIF occurrence data using clustering (i.e. DBSCAN) with haversine distance (sphere-earth distance).
It was inspired by this blog post: https://www.oreilly.com/content/clustering-geolocated-data-using-spark-and-dbscan/
The end result will be a table of outliers (probably around 1M-300K depending on the settings). Running on all Animal, Plant, and Fungi species with <20K unique points takes around 1 hour. Running with <30K unique points takes around 1.5 hours. <40K fails with out of memory errors.
- There are 222 993 specieskeys with less than 20K unique lat-lon points
- There are only 2 283 specieskeys with greater than 20K unique lat-lon points
The default (hard-coded) is to run only with species with >30 unique occurrence points.
An outlier of course does not mean that the point is in any way bad or a mistake. It simply is a point that is not close to any other points, which might mean the user should probably look at these points more closely. Outliers can be mistakes but they can also be valuable observations from an undersampled region. There is no objective way from distance alone to tell the difference.
Run inside project home directory where the build.sbt is located.
sbt assembly
The assembly jar gbif-dbscan-outliers-assembly-0.1.jar should be run with 3 cmd-line arguments:
- max_count : the max number of unique lat lon points per species to run. Should be between 20K - 50K. Species with more occurrences are not run.
- epsilon : the distance parameter in km for the dbscan algorithm. 1500 - 1800 probably good values
- minPoints : the minimum number of points for a cluster. 3 - 5 probably good values
spark2-submit --num-executors 40 --executor-cores 5 --driver-memory 8g --driver-cores 4 --executor-memory 16g gbif-dbscan-outliers-assembly-0.1.jar 20000 1500 3
This will save a file in hdfs called dbscan_outliers.
The raw output, while pretty good, probably needs some extra filtering before usable.
For example, the species below has way too many "outliers" flagged. One fix for this would be to white-list species that have more than 5% of its unique points as outliers.
Other species might have many points sitting exactly on one location from many observations over time. One could take the unique eventdate count of each occurrence and again white-list those occurrence locations for that species. Probably one would NOT want to white-list points which are simply duplicates on the same day ( for example this outlier point ).
Also one might not want to flag species only known from 1 dataset, since it is very likely this species is not well studied and therefore the points we have are not be mistakes even if far apart from each other. Below is one way to process the results into something more usuable for filtering.
val sqlContext = new org.apache.spark.sql.SQLContext(sc)
import sqlContext.implicits._;
val df = spark.read.
option("sep", "\t").
option("header", "false").
option("inferSchema", "true").
csv("dbscan_outliers").
withColumnRenamed("_c0","specieskey_outlier").
withColumnRenamed("_c1","decimallatitude_outlier").
withColumnRenamed("_c2","decimallongitude_outlier")
val df_occ = sqlContext.sql("SELECT * FROM prod_h.occurrence").
filter($"hasgeospatialissues" === false).
filter($"basisofrecord" =!= "FOSSIL_SPECIMEN").
filter($"basisofrecord" =!= "UNKNOWN").
filter($"basisofrecord" =!= "LIVING_SPECIMEN").
filter($"kingdomkey" === 1 || $"kingdomkey" === 6 || $"kingdomkey" === 5).
filter($"decimallatitude".isNotNull).
filter($"decimallongitude".isNotNull).
select("specieskey","decimallatitude","decimallongitude","gbifid","basisofrecord","datasetkey","kingdom","class","kingdomkey","classkey","eventdate","datasetname")
val df_outliers = df.join(df_occ,
df_occ("specieskey") === df("specieskey_outlier") &&
df_occ("decimallatitude") === df("decimallatitude_outlier") &&
df_occ("decimallongitude") === df("decimallongitude_outlier"),"left").
drop("decimallatitude_outlier").
drop("decimallongitude_outlier").
drop("specieskey_outlier").
withColumn("date",to_date(from_unixtime($"eventdate" / 1000)))
val df_outliers = df.join(df_occ,
df_occ("specieskey") === df("specieskey_outlier") &&
df_occ("decimallatitude") === df("decimallatitude_outlier") &&
df_occ("decimallongitude") === df("decimallongitude_outlier"),"left").
drop("decimallatitude_outlier").
drop("decimallongitude_outlier").
drop("specieskey_outlier").
withColumn("date",to_date(from_unixtime($"eventdate" / 1000)))
val df_species_outliers_counts = df.groupBy("specieskey_outlier").
agg(countDistinct("decimallatitude_outlier","decimallongitude_outlier").alias("species_unique_outlier_count")).
withColumnRenamed("specieskey_outlier","specieskey")
val df_dataset_counts = df_occ.groupBy("datasetkey").
agg(count(lit(1)).alias("dataset_occ_count"))
val df_species_counts = df_occ.groupBy("specieskey").
agg(
count(lit(1)).alias("species_occ_count"),
countDistinct("decimallatitude","decimallongitude").alias("species_unique_occ_count"),
countDistinct("datasetkey").alias("species_datasetkey_count")
)
val df_export_1 = df_dataset_counts.join(df_outliers,"datasetkey")
val df_export_2 = df_species_outliers_counts.join(df_export_1,"specieskey")
val df_export = df_species_counts.join(df_export_2,"specieskey").
cache()
df_export.select("specieskey","species_unique_outlier_count","species_unique_occ_count").show()
df_export.filter($"specieskey" === 8503754).select("specieskey","species_unique_outlier_count","species_unique_occ_count").show()
import org.apache.spark.sql.SaveMode
import sys.process._
val save_table_name = "dbscan_outliers_export"
df_export.
write.format("csv").
option("sep", "\t").
option("header", "false").
mode(SaveMode.Overwrite).
save(save_table_name)
// export and copy file to right location
(s"hdfs dfs -ls")!
(s"rm " + save_table_name)!
(s"hdfs dfs -getmerge /user/jwaller/"+ save_table_name + " " + save_table_name)!
(s"head " + save_table_name)!
// val header = "1i " + "specieskey\tspecies_occ_count\tdatasetkey\tdataset_occ_count\tdecimallatitude\tdecimallongitude\tgbifid\tbasisofrecord\tkingdom\tclass\tkingdomkey\tclasskey\teventdate\tdatasetname\tdate"
val header = "1i " + df_export.columns.toSeq.mkString("""\t""")
Seq("sed","-i",header,save_table_name).!
(s"rm /mnt/auto/misc/download.gbif.org/custom_download/jwaller/" + save_table_name)!
(s"ls -lh /mnt/auto/misc/download.gbif.org/custom_download/jwaller/")!
(s"cp /home/jwaller/" + save_table_name + " /mnt/auto/misc/download.gbif.org/custom_download/jwaller/" + save_table_name)!
https://stackoverflow.com/questions/36090906/dbscan-on-spark-which-implementation
https://github.com/haifengl/smile/blob/master/core/src/main/java/smile/clustering/DBSCAN.java Uses kd tree might scale better