For the integration of Jena and neo4j we have implemented Jena graph interfaces using neo4j native API. Neo4Jena is a property graph model interface. It provides the mapping of RDF to property graph (Neo4J) using Jena API. The main work focuses on how RDF triple is converted to Neo4j graph and vice versa. After the successful loading of RDF in graph we retrieve the data using SPARQL. We made the following contributions:
- Firstly RDF triples (subject, predicate and object) are converted to Neo4j node and relationship.
- After the conversion, it store triples in Neo4j graph.
- After the successful loading of RDF triples in graph Neo4Jena retrieve the data using SPARQL. For this neo4j nodes and relationship is converted to RDF triples (subject, predicate and object).
- Neo4Jena has full support of SPARQL 1.1.
RDF triple consist of subject, predicate and object.
- Subjects in rdf triples are URIs or blank nodes.
- All predicates are URIs.
- Objects can be URIs, blank nodes or literal values.
When mapping RDF to Neo4j following points are taken in considertaion:
- Each neo4j node has a label that is either uri , literal or bnode.
- Nodes having label uri or bnode have one property
- uri
- Nodes having label literal have three properties
- value
- datatype
- lang
Follwing figure demostrats how uri and literals are modeled in Neo4j.
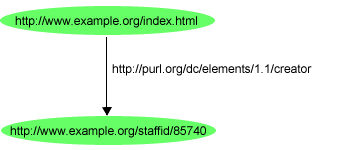
The following RDF statment is taken from RDF primer. This RDF statement have:
- a subject http://www.example.org/index.html
- a predicate http://purl.org/dc/elements/1.1/creator
- and an object http://www.example.org/staffid/85740
The subject and object are resources so while mapping them neo4j node has label uri.The following figure shows how this statement is represented in Neo4j. (In mapping prefixes are used)
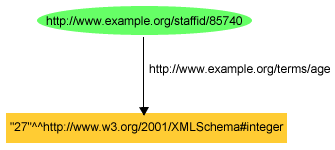
This RDF statement has:
- a subject http://www.example.org/staffid/85740
- a predicate http://example.org/terms/age
- and an object "27" which is a literal value.
The subject is a resource so while mapping them neo4j node has label uri.Literal values can have their datatype and language as well.
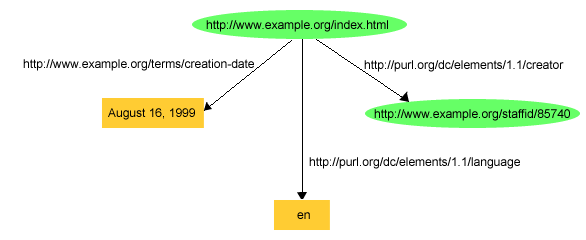
Following figure have multiple statements about the same subject. The object can be a URI or literal value.
Subjects and objects when mapped in neo4j node have label uri while literal values have label literal. The properties of uri and literals are different. !https://github.com/semr/neo4jena/raw/master/doc/image/example2.PNG!
Subject and object can also be a blank node (a resource having no URI)
While mapping blank node the neo4j node has label bnode.
!(alt tag
Create a Jena model and read RDF file/triples in it.
Model model = ModelFactory.createDefaultModel();
InputStream in = FileManager.get().open( inputFileName );
model.read(in,"","TTL"); For initialization of NeoGraph there are two constructors.
- public NeoGraph(final String directory)
- public NeoGraph(final GraphDatabaseService graphdb)
GraphDatabaseService njgraph = new GraphDatabaseFactory().newEmbeddedDatabase(NEO_STORE);
NeoGraph graph = new NeoGraph(njgraph);After initialization an instance of NeoGraph is created. Then create a Jena model for graph and pass NeoGraph instance as parameter.
Model njmodel = ModelFactory.createModelForGraph(graph);Load triples from model into njmodel.
njmodel.add(model);Create a Jena model and read RDF file/triples in it.
Model model = ModelFactory.createDefaultModel();
InputStream in = FileManager.get().open( inputFileName );
model.read(in,"","TTL"); Create a bacth inserter (act as graph service)
BatchInserter db = BatchInserters.inserter(NEO_STORE);Initialize a batch handler and register the model.
BatchHandler handler = new BatchHandler(inserter,500000,60);
model.register(handler);Read the model and close the handler
model.read(in,"","TTL");
handler.close();String queryString = "prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> \n" +
"PREFIX ub: <http://swat.cse.lehigh.edu/onto/univ-bench.owl#>" +
"SELECT ?X ?name "+
"WHERE" +
"{ ?X ub:name ?name ." +
"FILTER regex(?name,\"^Publication\") ."+
"}";
Query query = QueryFactory.create(queryString);
QueryExecution qExe = QueryExecutionFactory.create(query, njmodel);
ResultSet results = qExe.execSelect();