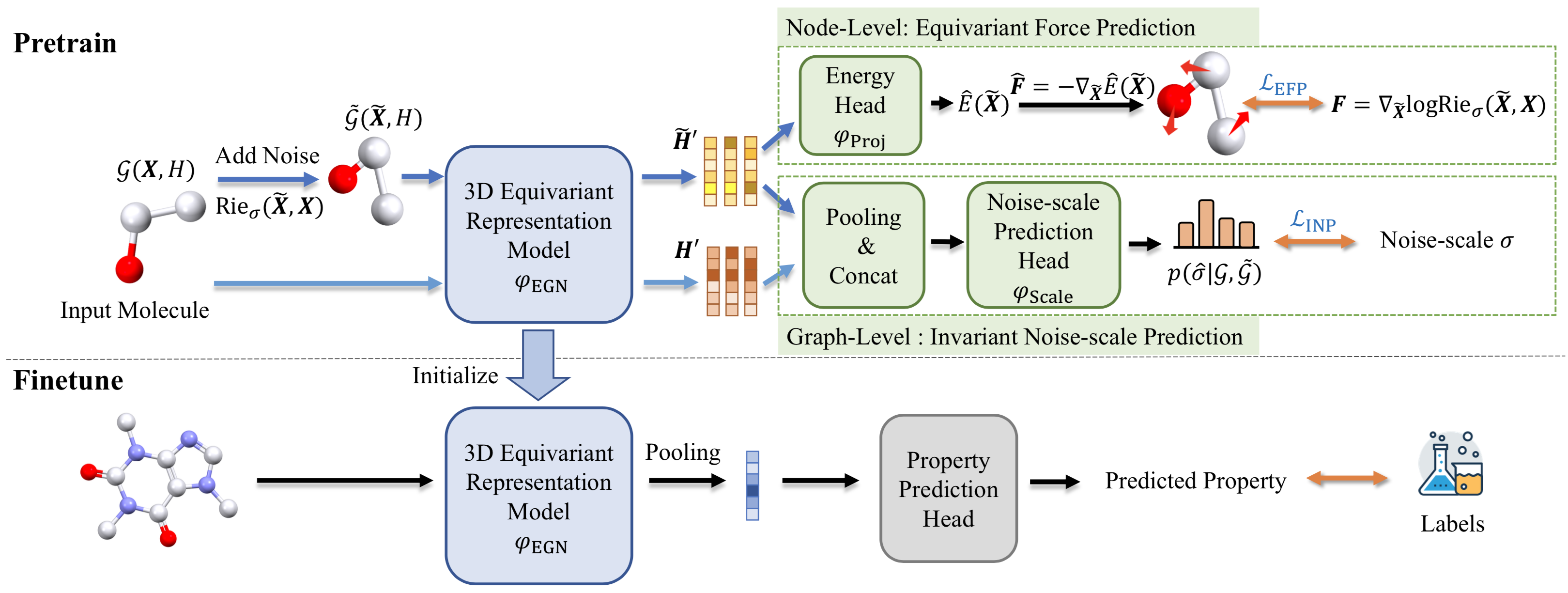
Code for Energy-Motivated Equivariant Pretraining for 3D Molecular Graphs (3D-EMGP).
python==3.7.10
torch==1.7.0
torch-geometric==1.6.3
The raw data of GEOM can be downloaded from the official website.
One can download the dataset and unpack it into the dataset folder as follows:
|-- datasets
|-- GEOM
|-- rdkit_folder
|-- drugs
|-- qm9
|-- summary_drugs.json
|-- summary_qm9.json
To preprocess the GEOM data for pretraining:
python data/geom.py --base_path datasets/GEOM/rdkit_folder/ --datasets qm9 --output blocks --val_num 500 --conf_num 10 --block_size 100000 --test_smiles data/filter_smiles.txt
Generated data blocks are listed as follows:
|-- datasets
|-- GEOM
|-- rdkit_folder
|-- blocks
|-- summary.json
|-- val_block.pkl
|-- train_block_i.pkl
3D-EMGP pretraining can be conducted via the following commands.
export CUDA_VISIBLE_DEVICES=0
python -u script/pretrain_3dmgp.py --config_path config/pretrain_3dmgp.ymlOne can also pretrain the model in a multi-GPU mode.
export CUDA_VISIBLE_DEVICES=0,1,2,3
python -m torch.distributed.launch --nproc_per_node=4 --master_port <port> script/pretrain_3dmgp.py --config_path config/pretrain_3dmgp.ymlThe pretrained model will be saved in checkpoints/pretrain/3dmgp, which can be modified in config/pretrain_3dmgp.yml
One can also pretrain the 3D model via the re-implemented baseline methods via the following commands. Take AttrMask as an example:
export CUDA_VISIBLE_DEVICES=0
python -u script/baselines/train_attr_mask.py --config_path config/pretrain_baselines.ymlFinetuning on QM9 :
python -u script/finetune_qm9.py --config_path config/finetune_qm9.yml --restore_path <pretrained_checkpoint> --property <property>
The property should be chosen from
alpha, gap, homo, lumo, mu, Cv, G, H, r2, U, U0, zpve
Finetuning on MD17 :
python -u script/finetune_md17.py --config_path config/finetune_md17.yml --restore_path <pretrained_checkpoint> --molecule <molecule> --model_name <molecule>
The molecule should be chosen from
aspirin
benzene
ethanol
malonaldehyde
naphthalene
salicylic_acid
toluene
uracil
Note that the finetuning datasets will be automatically downloaded and preprocessed on the first run.
Please consider citing our work if you find it helpful:
@misc{jiao2022energy,
url={https://arxiv.org/abs/2207.08824},
author={Jiao, Rui and Han, Jiaqi and Huang, Wenbing and Rong, Yu and Liu, Yang},
title={Energy-Motivated Equivariant Pretraining for 3D Molecular Graphs},
publisher={arXiv},
year={2022}
}