- Download executables for Linux, Mac OS
- For Mac-OS: follow the instruction to install Homebrew and then run "brew install zbar llvm", to install necessary libraries, and then double click "PENGUIN".
- For Linux: run "sudo apt-get install libzbar0 python3-pyqt5" to install packages, then run PAGEANT by typing "./PENGUIN".
- For advanced users, please check out the wiki for command line approaches.
Once the GUI window is displayed, a user simply needs to click the “Analyze” button to do a “test-drive” without doing anything else. Once a user gets familiar with the test-drive, he/she could change all parameters in the GUI interface:
- sample name
- genotype data (in VCF format)
- configuration file
- output location for the genetic report and log file
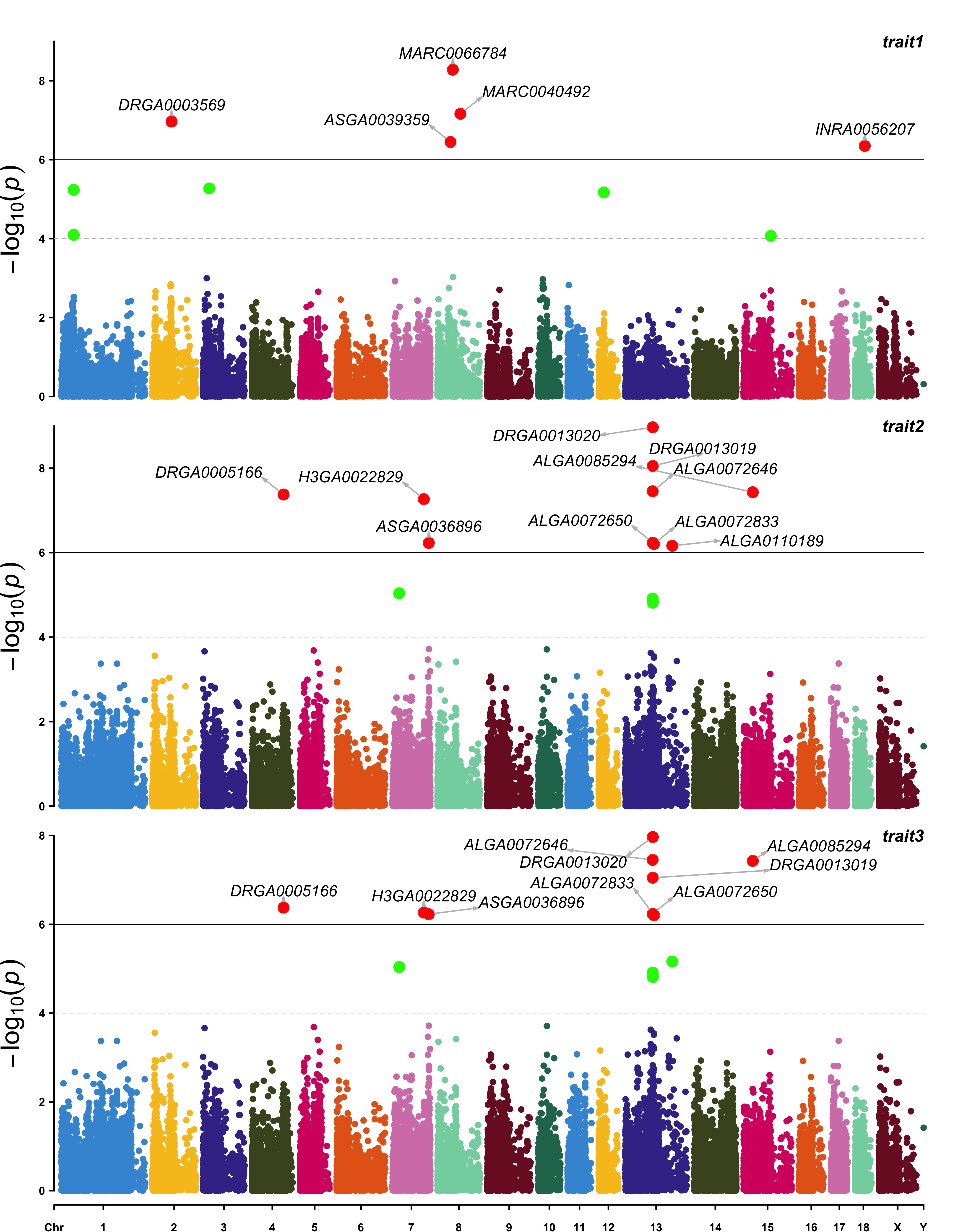
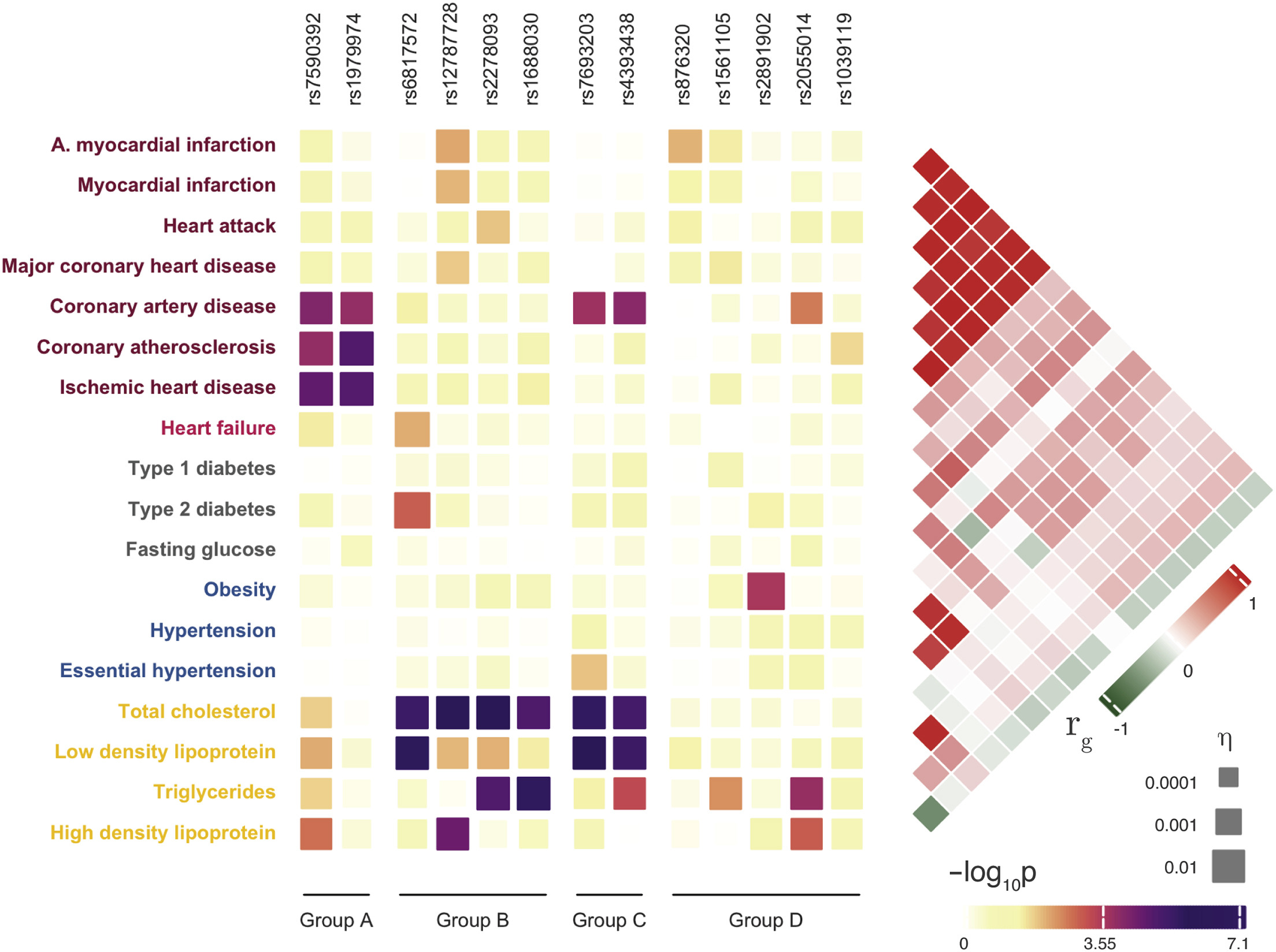
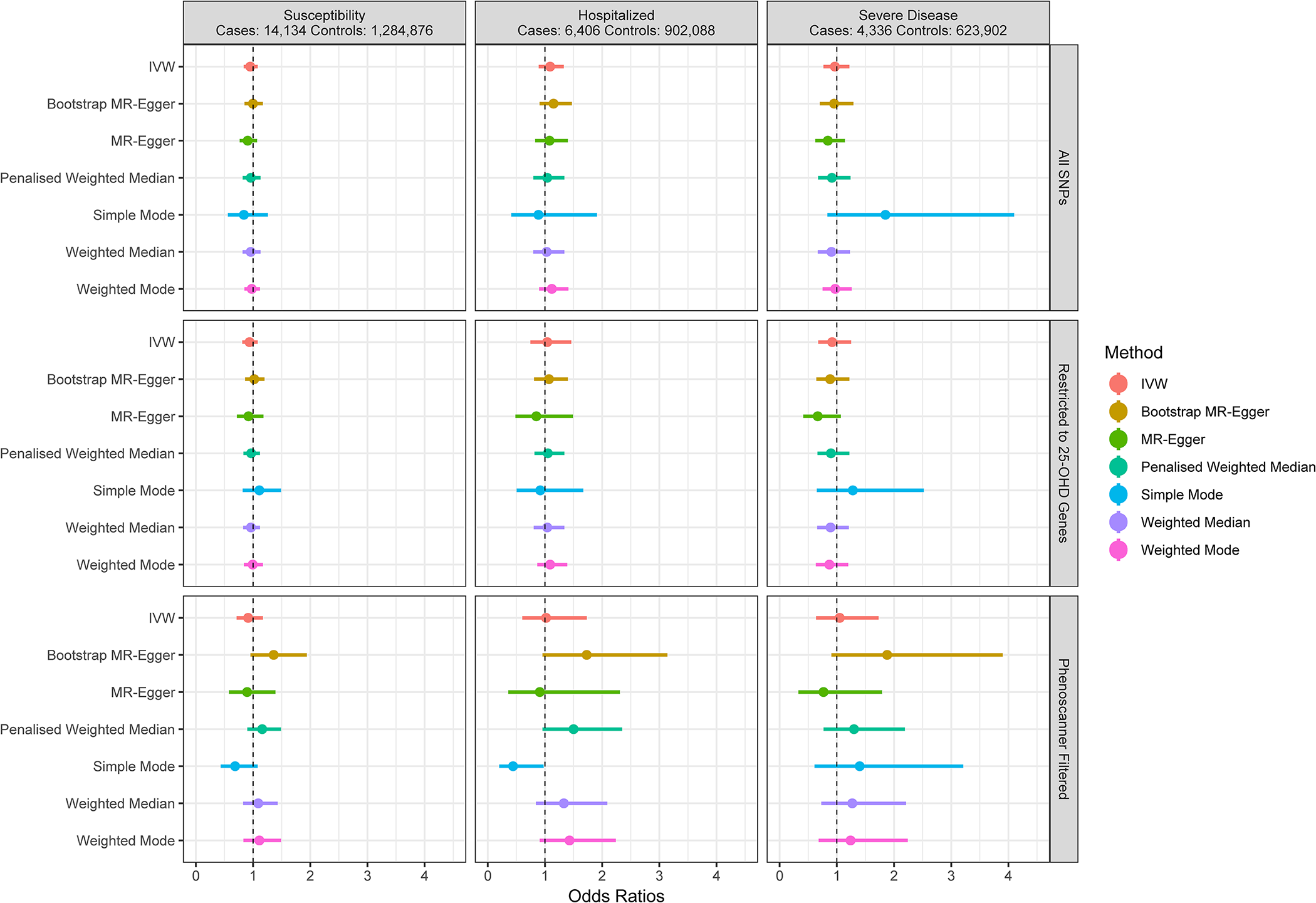
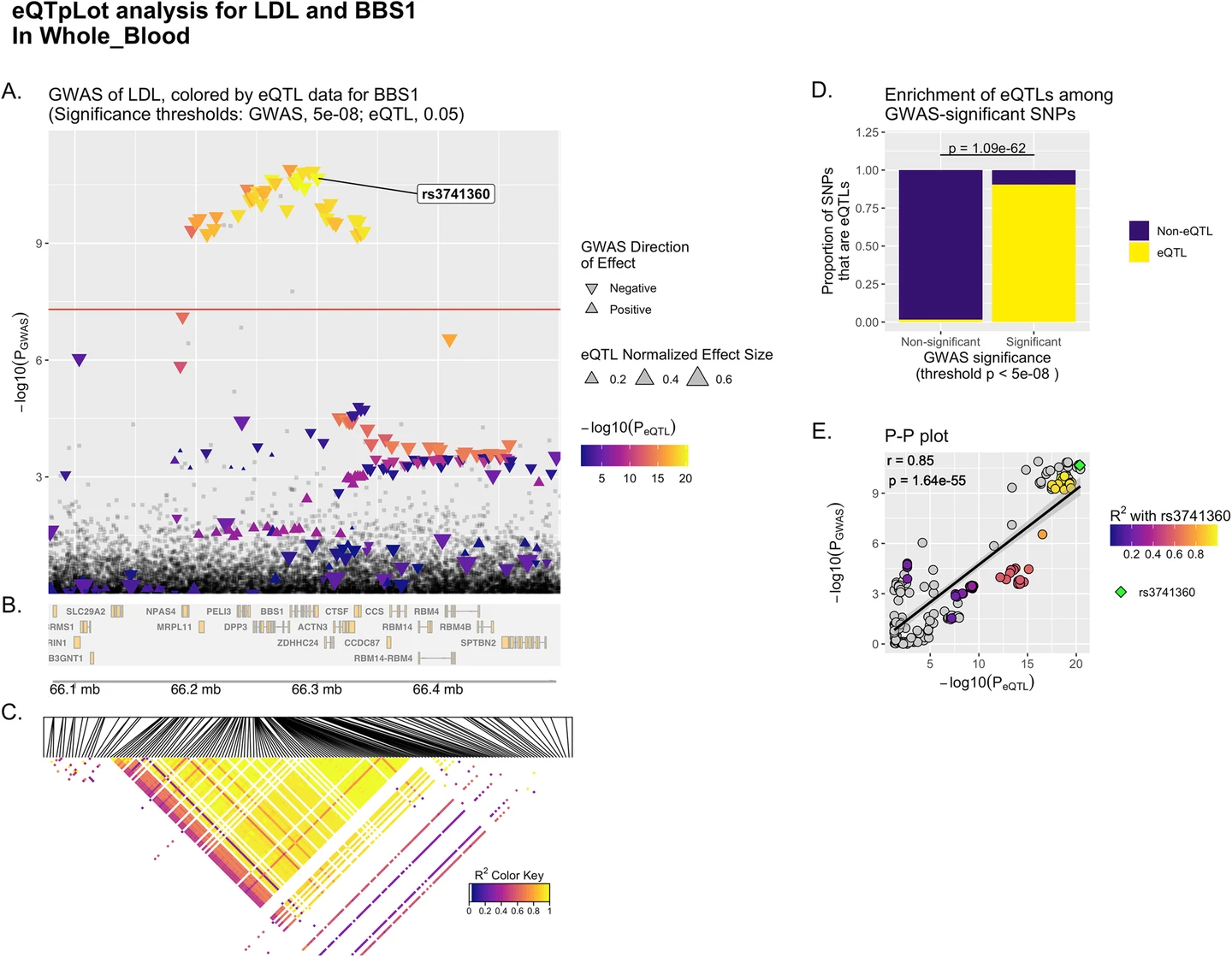
A full example genetic report can be viewed here.
- Jie Huang MD MPH PhD, Department of Global Health, Peking University School of Public Health
- Manuscript under review