FM Demo Repo
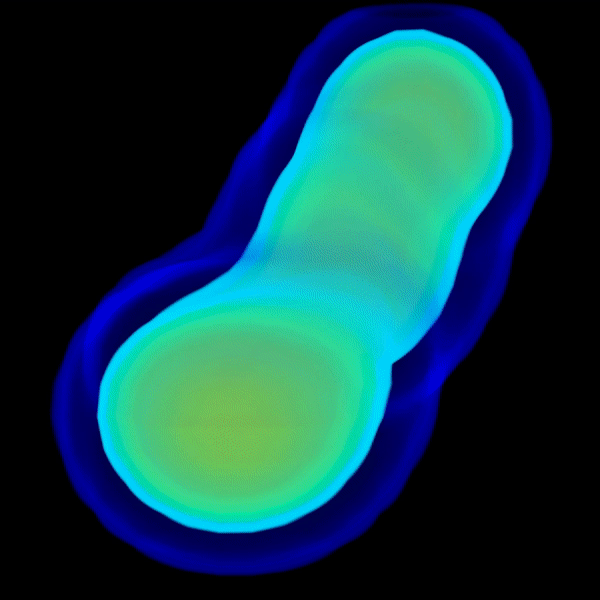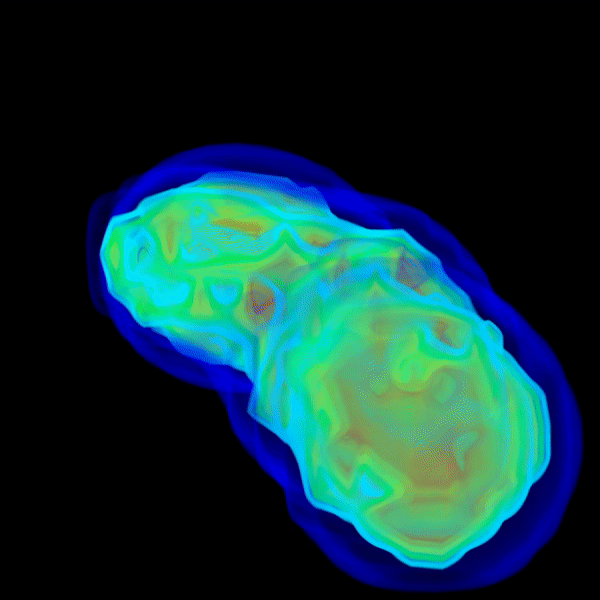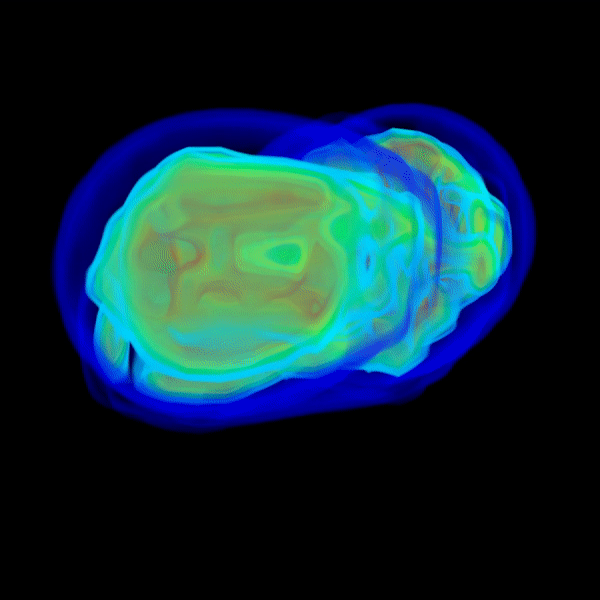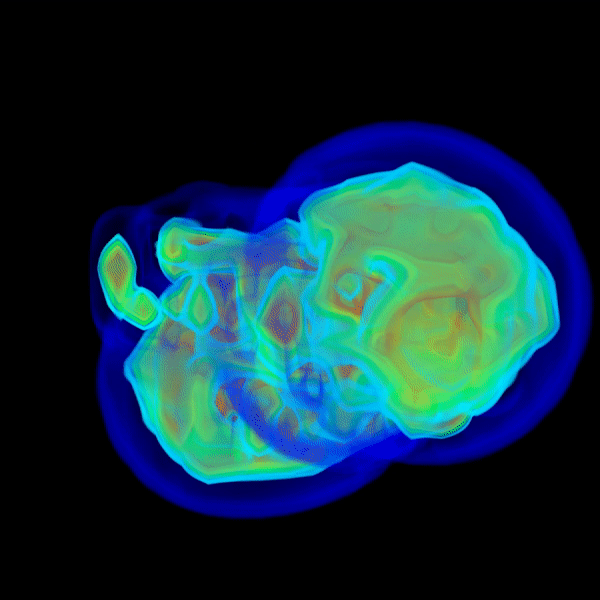
3D electron density reconstruction from solution X-ray scattering data into wide angles. It is suggested to run this on a multicore cluster or workstation because it utilizes julia's power of parallelization for speeding up.
- Mol 1
- Mol 2
- ArgParse.jl
- DelimitedFiles.jl
- LinearAlgebra.jl
- Statistics.jl
- Dierckx.jl
- PyCall.jl
- Conda.jl
- Reexport.jl
- SciPy
(Will be compiled with julia artifacts)
Run julia FrequencyMarching.jl --help
usage: freqmarching [-s STEPSIZE] [--f_tol F_TOL]
[--grad_tol GRAD_TOL] [-L LOWER] [-U UPPER]
[-M MAXITER] [--mvname MVNAME] [-p PERITER]
[-N FMSTEPS] [-d DECAY] [-c CUTOFF] [-G GRID]
[--qmin QMIN] [--qmax QMAX] [-Z ZOOM] [-J J]
[--qlead QLEAD] [--voxlead VOXLEAD] [-o OUTPUT]
[-n NPR] [-t] [-H] [-v] [-m] [--version] [-h]
priori input
Frequency Marching (FM) Algorithm for 3D electron density
reconstruction from solution X-ray scattering data into wide angles.
It is suggested to run on a multicore cluster or workstation because
it utilizes Julia's power of parallelization for speeding up.
positional arguments:
priori Reconstruction priori from DENSS, *.mrc
input Three-column experimental data, *.dat
optional arguments:
-s, --stepsize STEPSIZE
Step size, α, for ADAM. (type: Float64, default: 0.02)
--f_tol F_TOL Tolerance of χ^2 for ADAM. (type: Float64, default: 0.5)
--grad_tol GRAD_TOL Tolerance of |∇(χ^2)| for ADAM. (type: Float64, default: 0.08)
-L, --lower LOWER Lower bound of electron density, in e/A^3. (type: Float64, default: -0.01)
-U, --upper UPPER Upper bound of electron density, in e/A^3. (type: Float64, default: Inf)
-M, --maxiter MAXITER
Maximum iteration base, to be scaled based on FM steps. (type: Int64, default: 750)
--mvname MVNAME Directory where the movie frames are saved. (default: "mv_frame")
-p, --periter PERITER
Number of iteration gap bwteen 2 movie frames. (type: Int64, default: 10000)
-N, --fmsteps FMSTEPS
Number of Frequency Marching steps. (type: Int64, default: 6)
-d, --decay DECAY Gaussian Decay parameter. (type: Float64, default: 0.6)
-c, --cutoff CUTOFF Density cutoff of the input priori. (type: Float64, default: 0.001)
-G, --grid GRID Starting number of grids in each dimension. (type: Int64, default: 20)
--qmin QMIN Starting qmin for all Frequency Marching steps. (type: Float64, default: 0.02)
--qmax QMAX Starting qmax for the zero Frequency Marching step. (type: Float64, default: 0.25)
-Z, --zoom ZOOM Zoom parameter bridging between 2 Frequency Marching steps. (type: Float64, default: 1.25)
-J, --J J Number of 3D orientations, to be scaled based on FM steps. (type: Int64, default: 1200)
--qlead QLEAD (type: Float64, default: 1.12)
--voxlead VOXLEAD (type: Float64, default: 1.05)
-o, --output OUTPUT Output file prefix for saving the .dat file. (default: "fm_output")
-n, --npr NPR Number of parallel workers for computation. (type: Int64, default: 9)
-t, --trace Show real-time trace of each Frequency Marching step.
-H, --history Keep the statistics of each Frequency Marching step.
-v, --verbose Print out the statistics.
-m, --movie Save the movie frames of all Frequency Marching steps.
--version show version information and exit
-h, --help show this help message and exit
==============================================
Last updated: 01/13/20, Phnom Penh, Cambodia.
Copyright (c) Yen-Lin Chen, 2019 - 2020
Email: yc2253@cornell.edu
==============================================
Run the following in the command line
julia --color=yes -O 3 FrequencyMarching.jl -L -0.02 -M 500 --history dna_denss.mrc dna_swaxs.dat
You should see something like the following
Warming up Frequency Marching:
[ Info: Setting up parallel workers ...
========================================
┌ Warning: Frequency Marching : Data should include q=0 or scaling wwould be off ...
└ @ Main.FM C:\Users\Yen-Lin\Desktop\fmcomputation\fm\FM.jl:457
[ Info: Frequency Marching : FM will march to q = 1.882. Provided qmax = 1.99
[ Info: Frequency Marching : Back calculation from DENSS ...
[ Info: Frequency Marching : DENSS χ^2 = 56.87159124511503
[ Info: MRC: Operation completed (assigned) successfully ...
[ Info: MRC: Operation completed (assigned) successfully ...
[ Info: Frequency Marching : Starting χ^2 = 45.3105300505345,
[ Info: Frequency Marching : Setting up parameter scheduling ...
Starting Frequency Marching:
--------------------
[ Info: Frequency Marching : STEP=START=0, q range=[0.02, 0.25]
[ Info: Carve: The mrc size = (20, 20, 20), effective voxel number = 599 ...
[ Info: Initializing Adam Optimization (Full-batch Mode) With Constraints: Inf >= d >= -1.42...
[ Info: Adam Summary Of Iteration # 633 Under Full-Batch Mode:
====== χ^2 = 1.447; Elapsed Time = 0.4682
====== |grad| = 0.08; (max, min) = (0.008, -0.012)))
====== Total Density = 2430.512; (max, min) = (17.597, -0.524)
[ Info: Adam history has been saved as C:\Users\Yen-Lin\Desktop\fmcomputation\molecules\dna_a_denss1_fmstep0_history.dat ...
[ Info: MRC: The input ::mrc was modified (assigned) successfully ...
[ Info: MRC: C:\Users\Yen-Lin\Desktop\fmcomputation\molecules\dna_a_denss1_fmstep0_result.mrc written successfully!!
[ Info: MRC: C:\Users\Yen-Lin\Desktop\fmcomputation\molecules\dna_a_denss1_fmstep0_support.mrc written successfully!!
--------------------
And the above repeats as it marches into wider angles with greater resolutions until qmax is reached.
Using the script FreqMarching_cloud.jl. Put your .mrc and .dat file pair in a folder. If you have many file pairs, put all the folders in a directory, for example ../molecules/
Modify the pth
pth = "/path/to/molecules/";And run
julia --color=yes -O 3 FreqMarching_cloud.jl
It will run through all the data in the ../molecules/ directory. Execution time varies.
The following parameters are in the safe zone to play with.
--stepsize--f_tol--grad_tol--maxiter--qmin--qmax--J--output
The following parameters need more care to determine and is molecule-dependent.
--upper--lower
The rest of the parameters have been explored by the author to yield good results and performance. They are in the danger zone and it's recommended not to change those.
- The result depends on the priori
.mrc - For performance reasons, the
.datshould have increasingq-spacing. - The computation of the first 4 FM steps is fast, with
q < 1.0and the most computational effort is spent on FM steps that marches intoq > 1.0. - If everything is right, the solvent shell should show up by itself.
- The
q-voxel size relationship should be investigated more deeply. - Not sure if I will maintain this or not in the future ...