SWAXS computes the solution x-ray scattering profiles from orientational average of molecules in vitro with incorporation of the following theories:
- Spiral method for orientational average
- Debye Formula
- Theory of excessive electron densities
- Solvent/ion shells
- Excluded Volume
- Bulk solvent estimation (optional)
- Explicit solvent model or implicit solvent model
q-dependent atomic scattering form factor- Electron negativity in the
H-O-Hsolvent molecules
- This SWAXS program is written in Julia 1.6.0-DEV and built by PackageCompiler.jl
- SWAXS has 5 different computing modes for input files:
- Single
.pdbfile - A pair of
.pdbfiles:solute.pdbandsolvent.pdb(used for buffer subtraction and excluded volume) - 3D electron density map:
.mrcor.map. Buffer subtraction and excluded volume are implemented. - 3D shape with dummy voxels and customized uniform electron density:
.binvox. Buffer subtraction is implemented. - The
DAREmode with bulk solvent estimation from> 50explicit solvent frames:solute.pdband a directorybulkdircontainingprefix*.pdbfiles.
- By default, SWAXS uses all cores of the CPU and can be scaled for larger workstation and cluster.
Run .\bin\SWAXS --version
>.\bin\SWAXS --version
0.3.1
Run .\bin\SWAXS --help
>.\\bin\\SWAXS --help
usage: SWAXS [-D DENSITY] [-s SOLVENT_DENSITY] [-c DENSITY_CUTOFF]
[-P PDB] [-T SOLUTE] [-V SOLVENT] [-b BULKDIR]
[-e ENVELOPE] [-p PREFIX] [--dare] [-B BINVOX]
[-v VOXEL_DENSITY] [-J J] [-n NPR] [-o OUTPUT]
[--version] [-h] qmin qspacing qmax
Small and Wide-Angle X-ray Scattering calculator for (a) single atomic
coordinates: .pdb, (b) solute-solvent PDB pair for buffer subtraction,
(c) electron density: .mrc or .map with implicit density model, (d)
voxelized 3D shape: .binvox with uniform excessive electron density
and (f) DARE mode for accurate bulk solvent modeling. SWAXS implements
the Debye formula, orientational average and theory of excessive
electron density to account for buffer subtraction, solvent shell and
excluded volumes.
positional arguments:
qmin Starting q value, in (1/A) (type: Float64)
qspacing q grid spacing, in (1/A) (type: Float64)
qmax Ending q value, in (1/A) (type: Float64)
optional arguments:
-D, --density DENSITY
CCP4 electron density map file: .mrc or .map
-s, --solvent_density SOLVENT_DENSITY
The bulk solvent electron density in e/A^3
(type: Float64, default: 0.335)
-c, --density_cutoff DENSITY_CUTOFF
The electron density cutoff to exclude
near-zero voxels (type: Float64, default:
0.001)
-P, --pdb PDB Single file containing atomic coordinates:
.pdb
-T, --solute SOLUTE The solute.pdb file containing atomic
coordinates of molecules, ions and solvents
-V, --solvent SOLVENT
The solvent.pdb file containing atomic
coordinates of randomized bulk solvents
-b, --bulkdir BULKDIR
The directory containing bulk frames; more
than 50 frames are suggested or use --solute
and --solvent
-e, --envelope ENVELOPE
Distance between the envelope and molecular
surface, i.e. solvent layer width (type:
Float64, default: 10.0)
-p, --prefix PREFIX The .pdb file prefix in the --bulkdir
(default: "bulk")
--dare Make sure that you're actually doing it!!
Computing cluster suggested.
-B, --binvox BINVOX The shape file of dummy voxels: .binvox
-v, --voxel_density VOXEL_DENSITY
The averaged electron density on dummy voxels
in e/A^3 (type: Float64, default: 0.5)
-J, --J J Number of orientations to be averaged (type:
Int64, default: 1200)
-n, --npr NPR Number of parallel workers for computation
(type: Int64, default: 4)
-o, --output OUTPUT Output file prefix for saving the .dat file
(default: "output")
--version show version information and exit
-h, --help show this help message and exit
===================================================================
=== Last Update: 08/10/20, Ithaca, NY. ===
=== Copyright (c) Yen-Lin Chen, 2018 - 2020 ===
=== Academic Free License v. 3.0 ===
=== Email: yc2253@cornell.edu ===
===================================================================
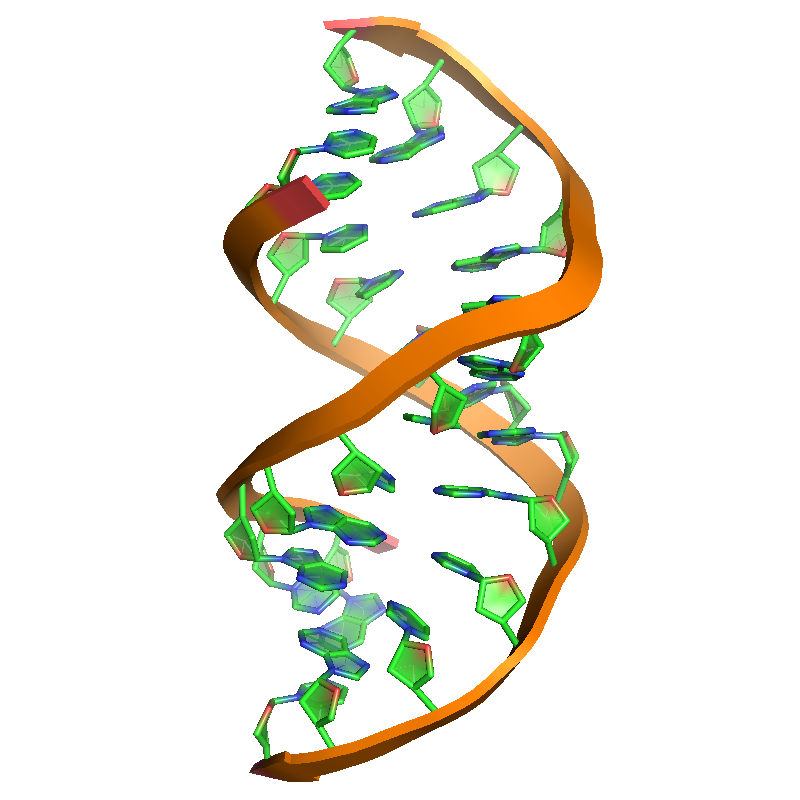
- The
.\test\rna.pdbfile contains about 750 atoms of short RNA duplex in vacuo. - Required argument(s):
--pdb
Run
.\bin\SWAXS --pdb ".\\test\\rna.pdb" -o test -J 1500 0.0 0.1 1.0
The output should look like
[ Info: --- SWAXS: Setting up parallel workers ...
[ Info: --- SWAXS: Please wait ...
[ Info: --- SWAXS: Computing SWAXS (J=1500) using single PDB file: .\\test\\rna.pdb.
[ Info: --- SWAXS: Starting Time: 2020-08-10T01:29:57.379 ...
[ Info: --- SWAXS: SWAXS program completed successfully: elapsed time = 17.63 seconds with 4 cores.
[ Info: --- SWAXS: Removing parallel workers ...
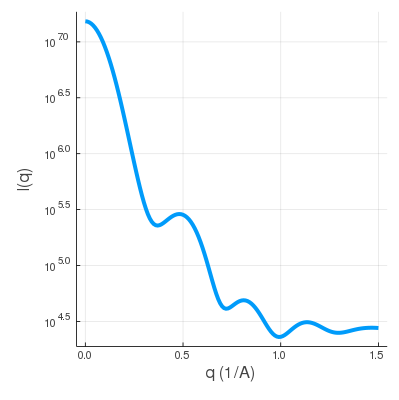
And the swaxs profile from q = 0.0 to q = 1.0 with spacing 0.1 is saved as test.dat.
| rna.pdb | SWAXS profile |
|---|---|
 |
 |
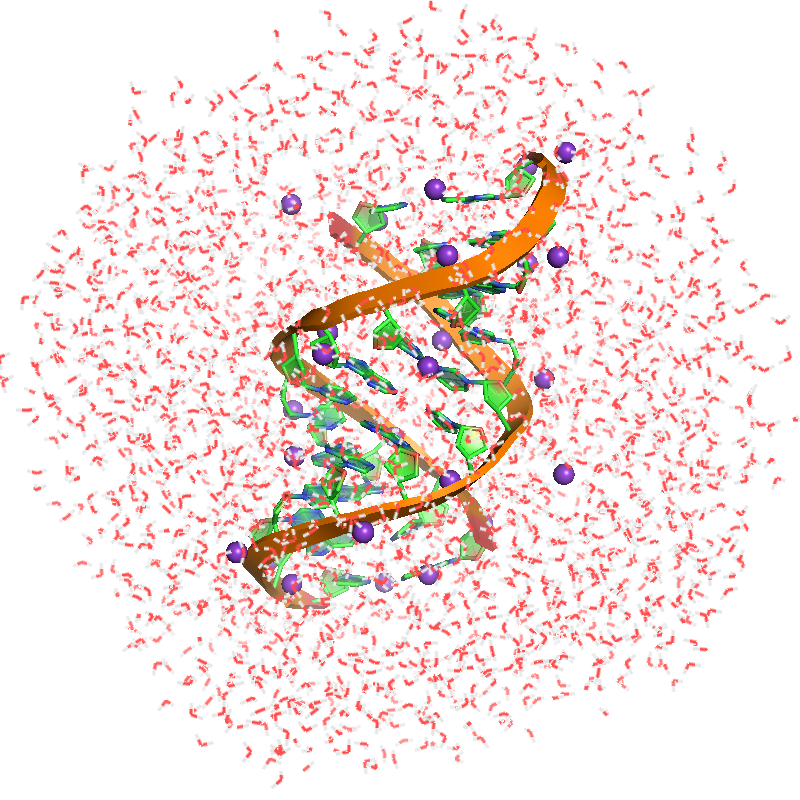
- Each of the
.pdbfile contains about 9000 atoms. Thesolvent.pdbis needed for buffer subtraction. - Required argument(s):
--solute,--solvent
Run
.\bin\SWAXS --solute ".\\test\\solute.pdb" --solvent ".\\test\\solvent.pdb" -o test 0.0 0.1 1.0
The output should look like
[ Info: --- SWAXS: Setting up parallel workers ...
[ Info: --- SWAXS: Please wait ...
[ Info: --- SWAXS: Processing solvent ...
[ Info: --- SWAXS: Computing SWAXS (J=1200) using solute: .\\test\\solute.pdb and solvent: .\\test\\solvent.pdb.
[ Info: --- SWAXS: Starting Time: 2020-08-10T01:38:43.683 ...
[ Info: --- SWAXS: SWAXS program completed successfully: elapsed time = 44.16 seconds with 4 cores.
[ Info: --- SWAXS: Removing parallel workers ...
And the swaxs profile from q = 0.0 to q = 1.0 with spacing 0.1 is saved as test.dat.
| solute.pdb | solvent.pdb | SWAXS profile |
|---|---|---|
 |
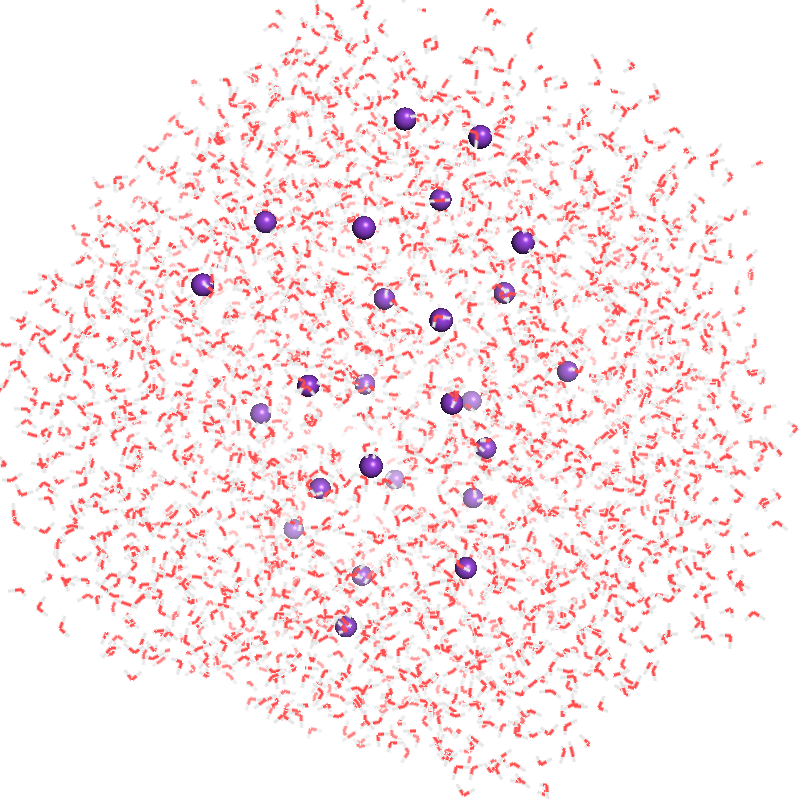 |
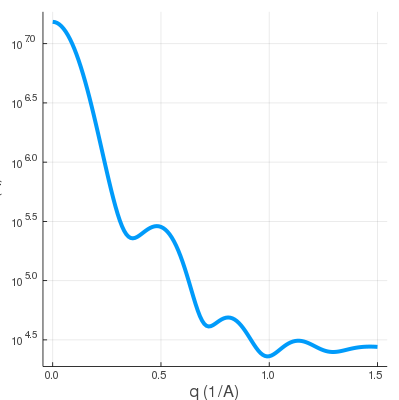 |
- The
dna1.mrccontains 96x96x96 volumetric data and is considered to be the excessive density on top of the uniform electron density in the solvent background. - Required argument(s):
--density,--solvent_density(the density for solvent background ine/A^3),--density_cutoff(the voxels beyond which are considered bulk-like)
Run
.\bin\SWAXS --density ".\\test\\dna1.mrc" -s 0.335 -c 0.001 -o test 0.0 0.1 1.0
The output should look like
[ Info: --- SWAXS: Setting up parallel workers ...
[ Info: --- SWAXS: Please wait ...
[ Info: --- SWAXS: Computing SWAXS (J=1200) using electron density file: .\\test\\dna1.mrc, with sden=0.335 cutoff=0.001.
[ Info: --- SWAXS: Starting Time: 2020-08-10T01:44:05.783 ...
[ Info: --- SWAXS: SWAXS program completed successfully: elapsed time = 24.28 seconds with 4 cores.
[ Info: --- SWAXS: Removing parallel workers ...
And the swaxs profile from q = 0.0 to q = 1.0 with spacing 0.1 is saved as test.dat.
| dna1.mrc | SWAXS profile |
|---|---|
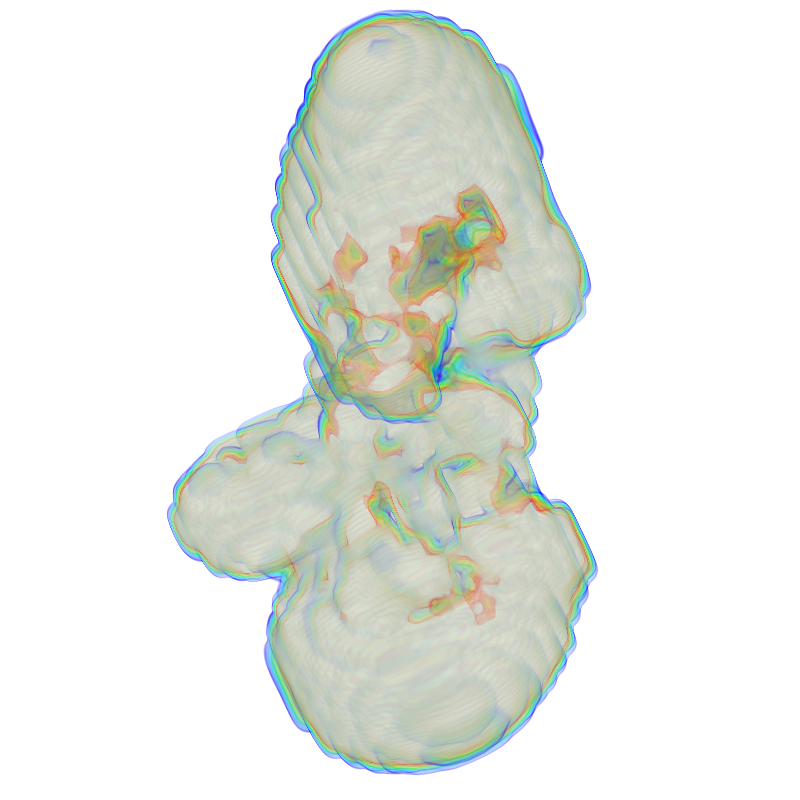 |
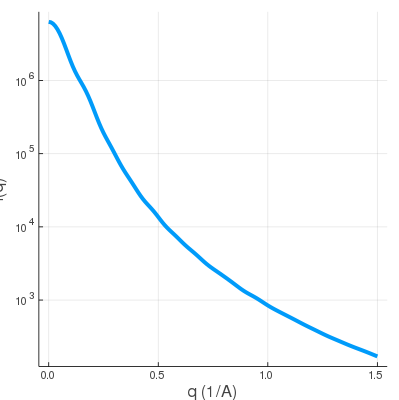 |
The dna1.mrc is just an envelope, so it doesn't have much feature in the WAXS regime.
| shape2.mrc | SWAXS profile |
|---|---|
 |
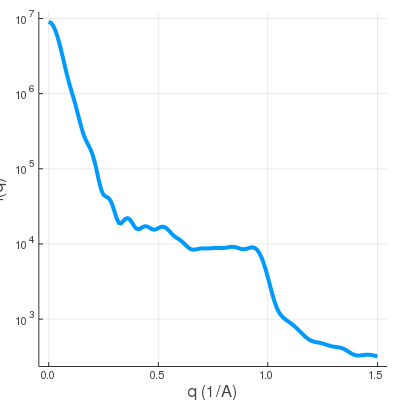 |
The shape2.mrc has more features, reflected in the wide-angle regime.
Note that this is just a demonstration and in this case, it's advised to have well-defined molecular support (not yet integrated in this SWAXS version.).
- The
rabbit.binvoxcontains 51x51x51 volumetric data with a grid size of2Aand is considered to be the 3D cat shape with Excessive Density1.0 e/A^3. - Required argument(s):
--binvox,--solvent_density(see above),--voxel_density(The uniform density on each voxel)
Run
.\bin\SWAXS --binvox ".\\test\\rabbit.binvox" -s 0.335 -v 1.0 -o test 0.0 0.1 1.0
The output should look like
[ Info: --- SWAXS: Setting up parallel workers ...
[ Info: --- SWAXS: Please wait ...
[ Info: --- SWAXS: Computing SWAXS (J=1200) using shape file: .\\test\\rabbit.binvox, with voxel_density=1.0, sden=0.335.
[ Info: --- SWAXS: Starting Time: 2020-08-10T01:50:47.430 ...
[ Info: --- SWAXS: SWAXS program completed successfully: elapsed time = 22.57 seconds with 4 cores.
[ Info: --- SWAXS: Removing parallel workers ...
And the swaxs profile from q = 0.0 to q = 1.0 with spacing 0.01 is saved as test.dat.
| rabbit.binvox | SWAXS profile |
|---|---|
 |
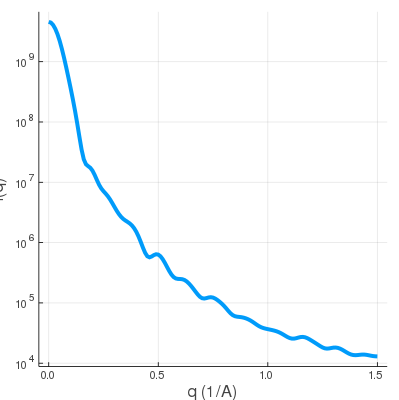 |
This uniform 3D voxelized shape is very similar to the shape.mrc case since the electron density is uniform within the rabbit. So SWAXS profile at wide-angle regime actually reveals finer periodicity from electron-denser structures.
- The
.\\test\\solventcontains the solute filesolute.pdband solvent frames from bulk MD simulationsframe*.pdbfor estimation of bulk solvent density. - In this case, all the solvent frames have the filename prefix of
frame. - More than 50 frames of random bulk solvent should be included.
- DARE mode requires computing cluster.
- Required argument(s):
--bulkdir,--prefix,--solute,--dare - The
--envelopeargument is optional with default of10.0 Ato include solvent and ion shells.
Run
.\bin\SWAXS --dare --bulkdir ".\\test\\solvent" --prefix frame --solute ".\\test\\solvent\\solute.pdb" --envelope 10.5 -J 800 -o test 0.0 0.1 1.0
The output should look like
[ Info: --- SWAXS: Setting up parallel workers ...
[ Info: --- SWAXS: DARE mode ...
[ Info: --- SWAXS: Processing bulk solvents ...
[ Info: -- Estimating solvent density using frame1.pdb ...
[ Info: -- Estimating solvent density using frame3.pdb ...
[ Info: -- Estimating solvent density using frame2.pdb ...
[ Info: --- SWAXS: Computing SWAXS (J=800) in DARE mode ...
[ Info: --- SWAXS: Starting Time: 2020-08-09T17:16:03.192 ...
[ Info: --- SWAXS: SWAXS program completed successfully: elapsed time = 483.1 seconds with 4 cores.
[ Info: --- SWAXS: Removing parallel workers ...
And the swaxs profile from q = 0.0 to q = 1.0 with spacing 0.1 is saved as test.dat.
- The options
--pdb,--density,--binvox,--solute --solventand--darecannot not be specified at the same time. Otherwise, error will be thrown. - If high-throughput computation is required, one should bypassing the command-line because it sets up parallel workers everytime. To avoid that, set up your parallel workers and call
@everywhere include("SWAXS.jl")and@everywhere using .SWAXSin the julia script. - The
--daremode is very expensive and should not be used for high-throughput computation unless on cluster. - The
--nprdefault option is machine-dependent. The following screenshot was taken on an 8-core PC (default = 8), showing the transferability ofSWAXSacross 32-bit and 64-bit Windows 10 machines.
- Every time
SWAXSis called, it creates--nprnumber of paralleljulias, even when only--helpor--versionis passed. This requires run-time ahead and might feel slow by just running.\bin\SWAXS --help. For checking the argument table and help, either refer to thisREADME.mdorREADME.pdfor try
.\bin\SWAXS --npr 1 --help
- Park, S. Simulated x-ray scattering of protein solutions using explicit-solvent models. The Journal of Chemical Physics 2009, 130, 134114
- Cromer, D.T. and Mann, J.B. X-ray scattering factors computed from numerical Hartree–Fock wave functions. Acta Crystallographica Section A 1968, 24, 321–324
- Sorenson, J.M., Hura, G., Glaeser, R.M. and Head-Gordon, T. What can x-ray scattering tell us about the radial distribution functions of water? The Journal of Chemical Physics 2000, 113, 9149–9161
- Ponti, A. Simulation of Magnetic Resonance Static Powder Lineshapes: A Quantitative Assessment of Spherical Codes. Journal of Magnetic Resonance 1999, 138, 288–297
- Chen, YL. et al., Salt Dependence of A-Form RNA Duplexes: Structures and Implications. J. Phys. Chem. B 2019, 123, 46, 9773-9785
- Chen, YL. et al., Machine learning deciphers structural features of RNA duplexes measured with solution X-ray scattering. IUCrJ 2020, 7, accepted
- He, W., Chen, YL. et al. in preparation
- Chen, YL. et al. On SARS-Cov-2 secondary structures. in preparation
- Chen, YL. et al. In vitro electron refinement. in preparation
Academic Free License v. 3.0
(c) Yen-Lin Chen, 2018 - 2020