We present here a novel group-wise graph learning method with the application in brain structural network. we assume that the observed brain network data consists of intrinsic connectivity information as well as external noise. To alleviate the issue of spurious connections at each brain network node, we introduce a sparsity constraint. To achieve the longitudinal consistency, we jointly consider the longitudinal sequence of networks in two ways. First, we examine the connectivity trajectory at each link of the network, where we deploy the kernel smoothing technique to prevent outlier connectivity due to the possible noise or computational error in the image processing pipeline. Second, we require high-level network geometry quantified by metrics such as network modularity to remain stable along time.
Our paper was accepted in MICCAI 2019 --- Link
We tested our method with the following environment
- python 3.6
- numpy 1.16.4
- matplotlib 3.1.0
- bctpy
- sklearn
Run test.py to run the method. This method takes input from data/sub_1 directory and writes the
intrinsic connectomes in data/sub_1_intrinsic directory. We have provided only one sample subject.
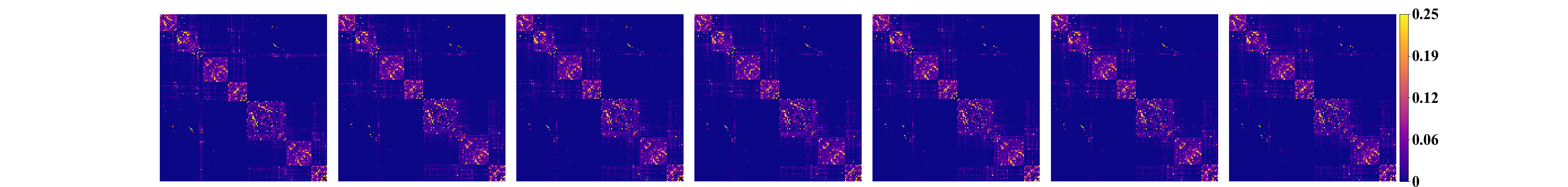
show_result.py generates the matrix plot for the raw and the intrinsic connectomes
and saves them in raw.png and intrinsic.png respectively.
Evaluation/evaluation_psnr.py contains the code for the first experiment.
eval_psnrplots the PSNR plot in Fig. 2(b).spurious_change_ratioprints the spurios change ratio mentioned in the paper
variation_of_information.py computes the NVI mentioned in the second experiment
in our paper.
classification/svm.py contains the code for the classification experiment which
is the third experiment in our paper. However, we haven't released all the data to
do the classification.
Md Asadullah Turja.
email: mturja@cs.unc.edu
Guorong Wu.
email: guorong_wu@med.unc.edu
Martin Styner.
email: styner@email.unc.edu