This repository is for maintaining the codes for RNA_seq dataset.
Download the Census of Immune Cells dataset from https://preview.data.humancellatlas.org.
This will create 4 folders with fastq.gz files. Unzip the files in each folder:
tar -xzf *.fastq.gz-
Download the cellranger sofware from here.
-
Download the GRCh38 as the reference.
-
Run cellranger count following the instructions from here. For example for MantonBM1:
cellranger count --id=MantonBM1 \
--fastqs=2a87dc5c-0c3c-4d91-a348-5d784ab48b92 \
--transcriptome=<path_to_reference_file> \
--sample=MantonBM1_HiSeq_1,MantonBM1_HiSeq_2,MantonBM1_HiSeq_3,MantonBM1_HiSeq_4,MantonBM1_HiSeq_5,MantonBM1_HiSeq_6,MantonBM1_HiSeq_7,MantonBM1_HiSeq_8This will create a folder MantonBM1.
Locate the filtered_gene_bc_matrices_h5.h5 file.
You have following parameters to tune for different setting:
- cutoff_thresh : Remove genes that the sum of counts is below threshold
- dim_red_method: Dimension reduction method { 'PCA' , 'SNE'}
- red_dim : Reduced dimension
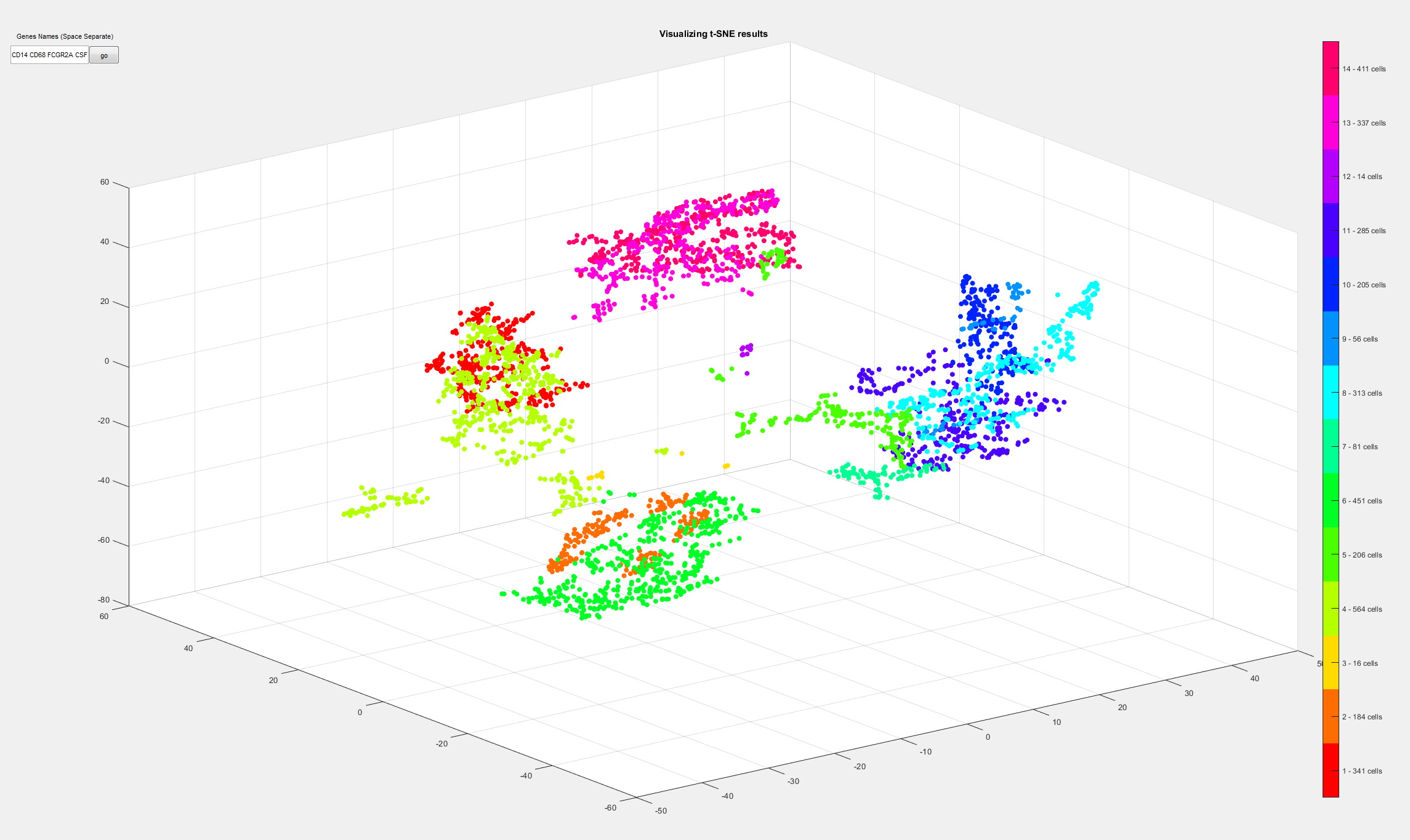
It will visualize samples in a color-coded manner like this:
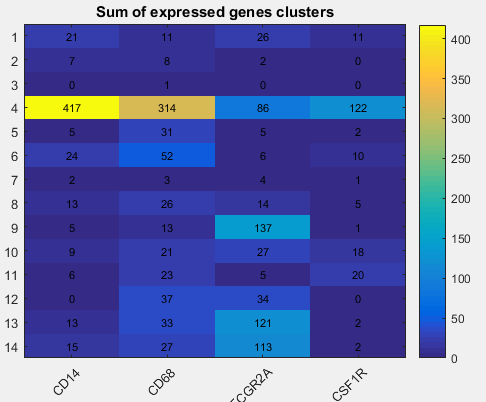
You can provide gene names (space separate the names) and see the heatmap of the sum of expressesd genes in each cluster.