[IEEE TMI 2024] WIN: A simple normalization technique using window statistics to improve the out-of-distribution generalization on medical images
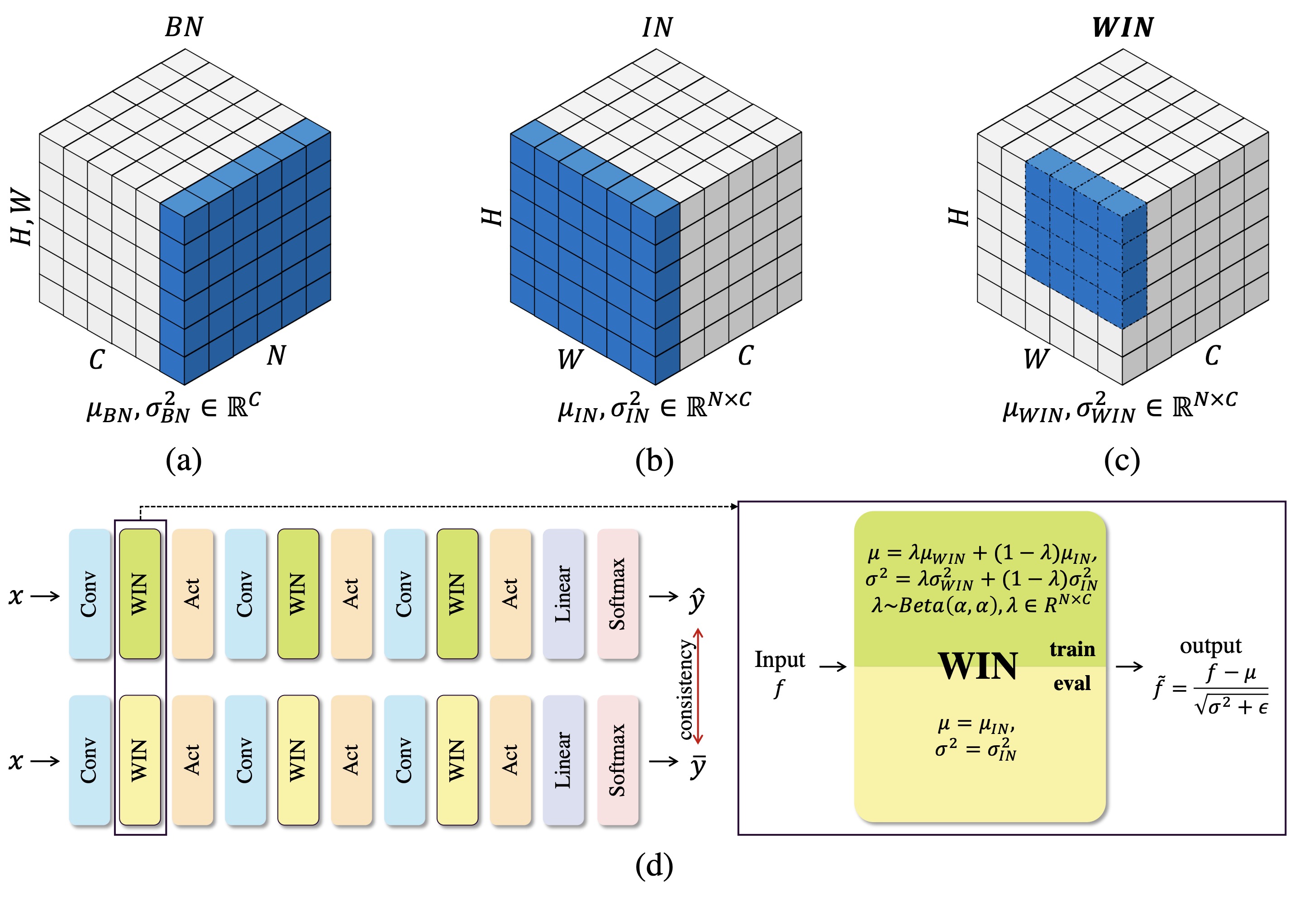
Since data scarcity and data heterogeneity are prevailing for medical images, well-trained Convolutional Neural Networks (CNNs) using previous normalization methods may perform poorly when deployed to a new site. However, a reliable model for real-world clinical applications should be able to generalize well both on in-distribution (IND) and out-of-distribution (OOD) data (e.g., the new site data). In this study, we present a novel normalization technique called window normalization (WIN) to improve the model generalization on heterogeneous medical images, which is a simple yet effective alternative to existing normalization methods. Specifically, WIN perturbs the normalizing statistics with the local statistics computed on the window of features. This feature-level augmentation technique regularizes the models well and improves their OOD generalization significantly. Taking its advantage, we propose a novel self-distillation method called WIN-WIN for classification tasks. WIN-WIN is easily implemented with twice forward passes and a consistency constraint, which can be a simple extension for existing methods. Extensive experimental results on various tasks (6 tasks) and datasets (24 datasets) demonstrate the generality and effectiveness of our methods.
Read the paper here.
The precomputed windows may be enabled for faster training by setting the variable cached=False.
They are available here.
- numpy==1.19.5
- Pillow==8.3.2
- torch==1.10.0
- torchvision==0.11.1
Download CIFAR-10-C and CIFAR-100-C datasets with:
mkdir -p ./data/cifar
curl -O https://zenodo.org/record/2535967/files/CIFAR-10-C.tar
curl -O https://zenodo.org/record/3555552/files/CIFAR-100-C.tar
tar -xvf CIFAR-100-C.tar -C data/cifar/
tar -xvf CIFAR-10-C.tar -C data/cifar/
Training recipes used in our paper:
WIN:
python cifar.py --dataset cifar10 --norm WIN
python cifar.py --dataset cifar100 --norm WIN
WIN-WIN
python cifar.py --dataset cifar10 --norm WIN-WIN
python cifar.py --dataset cifar100 --norm WIN-WIN
You can use this following helper function to convert all torch.nn.BatchNorm2d layers in the model to WindowNorm2d layers.
net = WindowNorm2d.convert_WIN_model(net)In addition, we offer two helper functions, convert_GN_model and convert_IN_model, that enable the conversion of all torch.nn.BatchNorm2d layers in the model to their respective layers.
Takeaways on how to apply WindowNorm2d to your tasks:
mask_thresis the most important hyper-parameter, which empirical set to 0.3~0.7.- Block strategy (i.e.,
grid=True) is more suitable for the image with a consistent background. - Advanced data augmentation techniques, such as AutoAug and RandAug, do not provide significant improvements.
| Normalization | CIFAR-10 (Acc.) | CIFAR-10-C (mCE) | CIFAR-100 (Acc. ) | CIFAR-100-C (mCE) |
|---|---|---|---|---|
| BatchNorm | 94.0±0.2 | 25.8±0.3 | 74.8±0.2 | 51.5±0.7 |
| GroupNorm | 91.2±1.2 | 23.6±1.8 | 66.1±0.9 | 55.5±0.5 |
| IN | 94.4±0.1 | 18.4±0.3 | 74.4±0.3 | 48.7±0.6 |
| WIN | 94.1±0.1 | 18.3±0.3 | 74.7±0.2 | 46.7±0.4 |
Weights for a ResNet-18 CIFAR-10 classifier trained with WIN for 180 epochs are available here.
Weights for a ResNet-18 CIFAR-100 classifier trained with WIN for 200 epochs are available here.
joe1chief1993 at gmail.com
Any discussions, suggestions and questions are welcome!
To cite WIN in your publications, please use the following bibtex entry
@article{zhou2024simple,
title={A simple normalization technique using window statistics to improve the out-of-distribution generalization on medical images},
author={Zhou, Chengfeng and Wang, Jun and Xiang, Suncheng and Liu, Feng and Huang, Hefeng and Qian, Dahong},
journal={IEEE Transactions on Medical Imaging},
year={2024},
publisher={IEEE}
}