The LabRecorder is the default recording program that comes with LSL. It allows to record all streams on the lab network (or a subset) into a single file, with time synchronization between streams.
The file format used by the LabRecorder is XDF. This is an open general-purpose format that was designed concurrently with LSL and supports all features of LSL streams. The project page is here. There are importers for MATLAB, EEGLAB, BCILAB, Python, and MoBILAB.
For LabRecorder to work on your system, you might need to first install some dependencies.
In the near future it will be necessary to use homebrew to manage LSL Apps and their dependencies:
- Install homebrew:
/bin/bash -c "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/HEAD/install.sh)"
The Ubuntu releases do not typically ship with their dependencies so you will also have to download and install those:
- Download and install the latest liblsl-{version}-bionic_amd64.deb from its release page
- We hope to make this available via a package manager soon.
- You can install liblsl directly by double clicking on it, or with with
sudo dpkg -i {filename}.deb
- See the bottom of the lsl build env docs.
- For most cases, this will amount to
sudo apt-get install qtbase5-dev
- For most cases, this will amount to
-
brew install labstreaminglayer/tap/labrecorder -
Run it with
open /usr/local/opt/labrecorder/LabRecorder/LabRecorder.app
Navigate to the releases page and download the latest release for your platform.
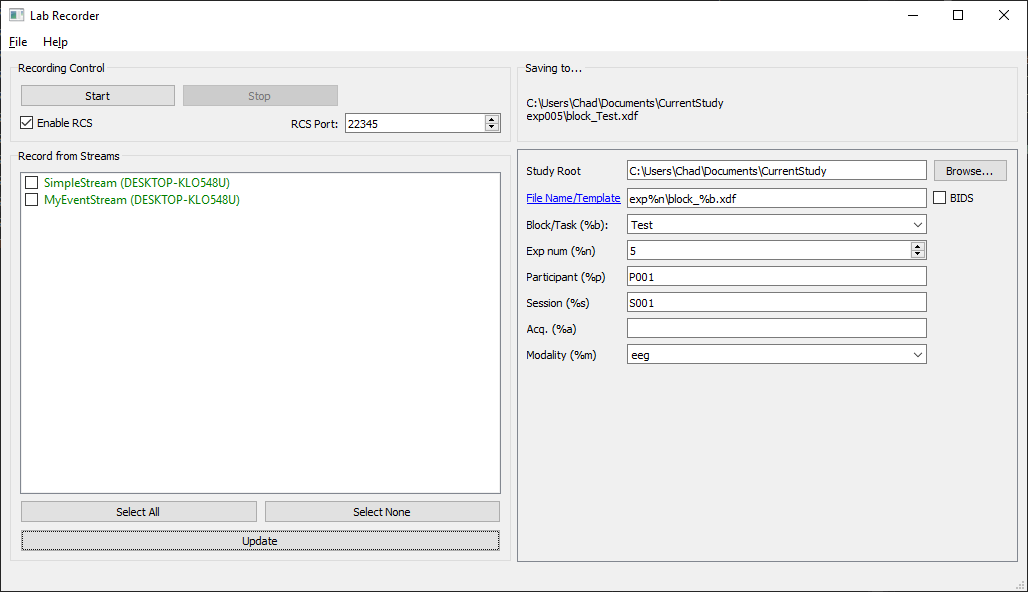
The LabRecorder displays a list of currently present device streams under "Record from Streams". If you have turned on a device after you have already started the recorder, click the "Update" button to update the list (this takes ca. 2 seconds).
For testing you can use a "dummy" device from the
lslexamplesfound in the liblsl release assets (for example SendData).
If you cannot see streams that are provided on another computer, read the section Network Troubleshooting on the NetworkConnectivity page.
You can select which streams you want to record from and which not by checking the check boxes next to them.
Note that if you have multiple streams with the same name and host, it is impossible to check only 1. If any is checked then they will all be recorded.
The entry in "Saving to..." shows you the file name (or file name template) where your recording will be stored. You can change this by modifying the Study Root folder (e.g., by clicking the browse button) and the File Name / Template field. If the respective directory does not yet exist, it will be created automatically (except if you do not have the permissions to create it). The file name string may contain placeholders that will be replaced by the values in the fields below. Checking the BIDS box will automatically change the filename template to be BIDS compliant. If the file that you are trying to record to already exists, the existing file will be renamed (the string _oldX will be appended where X is the lowest number that is not yet occupied by another existing file). This way, it is impossible to accidentally overwrite data.
The Block/Task field can be overwriten or selected among a list of items found in the configuration file.
Click "Start" to start a recording. If everything goes well, the status bar will now display the time since you started the recording, and more importantly, the current file size (the number before the kb) will grow slowly. This is a way to check whether you are still in fact recording data. The recording program cannot be closed while you are recording (as a safety measure).
When you are done recording, click the "Stop" button. You can now close the program. See the xdf repository for tools and information on how to use the XDF files.
When preparing a new study, it is a good idea to make a custom configuration file which at least sets up a custom storage location for the study. See the documentation in the file LabRecorder.cfg for how to do this -- it is very easy! You can override this by making a shortcut for the LabRecorder program (e.g. on the desktop) and appending in its properties the command-line arguments -c name_of_you_config.cfg. You can also create a batch script. You can also load the config while the program is already running, but this can easily be forgotten during an experiment, so we recommend to follow the shortcut route.
In addition to the storage location, if your experiment has multiple blocks (e.g., SubjectTraining, PreBaseline, MainBlock, PostBaseline or the like) you can make the recording process even more straightforward for the experimenters by setting up a default list of block names. Again, take a look at the existing config files.
Since it is too easy to forget to turn on or check all necessary recording devices for a study, we recommend to also make a list of "required" streams (by their name) and put it into the config file. These streams will be pre-checked when starting the program, and any missing stream will be displayed in red. If such a stream is still not green when starting the recording, the experimenter will get a message box to confirm that he/she really wants to record without including the device.
If you check the box to EnableRCS then LabRecorder exposes some rudimentary controls via TCP socket.
Currently supported commands include:
select allselect nonestartstopupdatefilename ...
filename is followed by a series of space-delimited options enclosed in curly braces. e.g. {root:C:\root_data_dir}
root- Sets the root data directory.template- sets the File Name / Template. Will unselect BIDS option. May contain wildcards.task- will replace %b in templaterun- will replace %n in template (not working?)participant- will replace %p in templatesession- will replace %s in templateacquisition- will replace %a in templatemodality- will replace %m in template. suggested values: eeg, ieeg, meg, beh
For example, in Python:
import socket
s = socket.create_connection(("localhost", 22345))
s.sendall(b"select all\n")
s.sendall(b"filename {root:C:\\Data\\} {template:exp%n\\%p_block_%b.xdf} {run:2} {participant:P003} {task:MemoryGuided}\n")
s.sendall(b"start\n")lr = tcpip('localhost', 22345);
fopen(lr)
fprintf(lr, 'select all');
fprintf(lr, ['filename {root:C:\Data\} '...
'{task:MemoryGuided} ' ...
'{template:s_%p_%n.xdf ' ...
'{modality:ieeg}']);
fprintf(lr, 'start');The LabRecorder has some useful features that can add robustness if things go wrong during the experiment:
If a network connectivity error happens while recording (e.g., a network cable pops out that connects to the source of a data stream), you have usually 6 minutes (think 5) to plug it back it in during which the data will be buffered on the sending machine. If it takes you longer to fix the problem, you will have some gap in your recording.
If a device program or computer crashes while recording, you will for sure lose data, but any device program that transmits an associated device serial number will be picked up automatically by the recorder when it comes back online (these programs are called "recoverable"), without a need to stop and re-start the recording.
You should check the health of your device to be sure, however, for example using an online stream viewer program (see, for example, ViewingStreamsInMatlab). Also, be sure to test whether it is in fact recoverable before relying on this feature (you can test this with a viewer by turning the device app off and back on).
If a device is displayed in red when you start recording (and it is checked), it will be added to the ongoing recording by the time when it comes online. This can be useful when a device can only be turned on while the recording is already in progress. Again, it is advisable to check that the device is in fact discoverable and added. The LabRecorder brings up a console window in the background which shows a list of all streams that are added to the recording -- this is a good place to check whether a late stream did get picked up successfully during a live recording.
Please follow the general LSL App build instructions.