A tool to crawl machine learning proceedings for abstracts, pdfs, key words, bibtex et cetera and populate a database with these. Then match the abstracts of the papers in the database against one of your own abstracts and rank by similarity, and display them in a simple interface that supports opening the papers in the browser, and copying their bibtex entries to the clipboard for proper citing.
pip install -r requirements.txt
- beautifulsoup4
If you want to use abstract similarity script also:
- transformes
- torch
python3 src/crawl.py --venue=arxiv --query_term='"noisy labels"' --database=databases/noisy_labels.db
This will crawl 'arxiv' for papers with "active learning" in the title and insert them into databases/papers.db. The query functionality is limited (see def matches_query in parse_site.py), but just keep to lower case characters and use the same format as above and it should be fine.
supported venues:
- iclr, back to 2018,
- iclm, back to 2013,
- neurips, back to 1988
- arxiv
ICLR constrains the number of queries, so this takes time if you have many hits. It is also a bit buggy still and may crash.
Run the
bash example_crawl.sh
script for a crawl over a list of query terms and venues. This will crawl for machine learning papers on learning from noisy labels and populate a paper database 'databases/noisy_labels.db with these.
Run the script
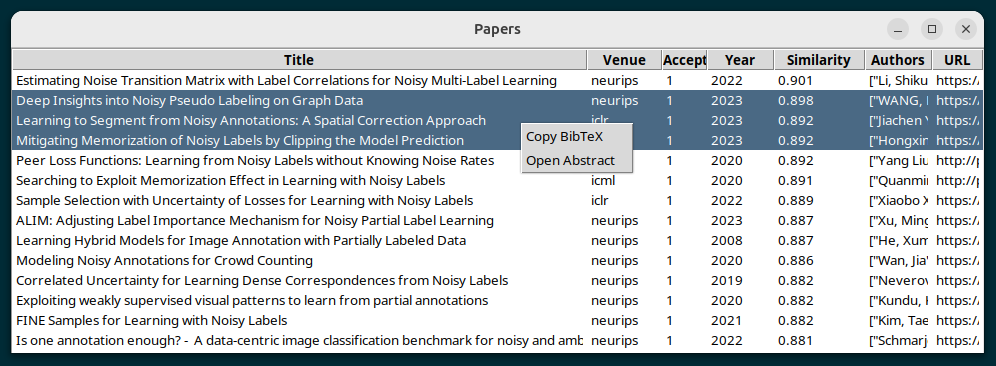
python src/display_papers.py databases/noisy_labels.db
to display the papers.
Functionality: - double left-click : open paper in browser - single right-click : open context menu - open abstract - copy bibtex
Many papers can be selected by CTRL or SHIFT selecting, and then all abstracts of the selected papers will be open, and all bibtex entries of the selected papers will be copied.
If this has not been run the 'similarity' column when displaying papers will be '0'. However, if you want to sort papers according to abstract similarity to a pre-defined weighted sentence list using transformers, then run this command:
python src/compute_similarities.py --database=databases/noisy_labels.db --random_papers=0 --sentence_list_name=noisy_labels
This will rank all the paper abstracts with the predefined weighted sentences in the sentence list "noisy_labels" in compute_similarities.py, change to your liking to get relevant similarity scores. If you set --random_papers > 0 a random selection of all papers will be chosen for the ranking. Can be useful when building a new sentence list to iterate quickly and get a feeling for what type of matches it produces.
Next time you display the papers you can sort by this similarity.