Fast implementation of HMMSearch optimized for high-memory systems using PyHmmer. PyHMMSearch can handle fasta in uncompressed or gzip format and databases in either HMM or Python pickle serialized format. No intermediate files are created.
pip install pyhmmsearch
- pyhmmer >=0.10.12
- pandas
- tqdm
| Database | Tool | Single Threaded | 12 Threads |
|---|---|---|---|
| Pfam | PyHMMSearch | 2:24 | 0:20 |
| Pfam | HMMER HMMSearch | 2:53 | 2:27 |
* Time in minutes for 4977 proteins in test/test.faa.gz.
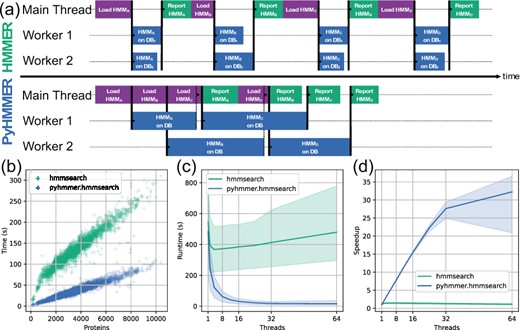
Official benchmarking for hmmsearch algorithm implemented in PyHMMER against HMMER from Larralde et al. 2023:
Recommended usage for PyHMMSearch is on systems with 1) high RAM; 2) large numbers of threads; and/or 3) reading/writing to disk is charged (e.g., AWS EFS). Also useful when querying a large number of proteins.
-
# Download database DATABASE_DIRECTORY=/path/to/database_directory/ mkdir -p ${DATABASE_DIRECTORY}/Annotate/Pfam wget -v -P ${DATABASE_DIRECTORY}/Annotate/Pfam https://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz # Run PyHMMSearch pyhmmsearch -i test/test.faa.gz -o output.tsv -b ${DATABASE_DIRECTORY}/Annotate/Pfam/Pfam-A.hmm.gz -p=-1
-
# Provide a database serialize_hmm_models -d path/to/Pfam-A.hmm.gz -b path/to/database.pkl.gz # or a directory of HMMs serialize_hmm_models -d path/to/hmm_directory/ -b path/to/database.pkl.gz # or from a list of filepaths to HMM models serialize_hmm_models -l path/to/hmms.list -b path/to/database.pkl.gz # or form a list through stdin ls path/to/directory/*.hmm | serialize_hmm_models -b path/to/database.pkl.gz
-
Database can be uncompressed pickle or gzipped pickle.
pyhmmsearch -i test/test.faa.gz -o output.tsv -b ~/Databases/Pfam/database.pkl.gz -p=-1 -
pyhmmsearch -i test/test.faa.gz -o output.tsv -d test/bacteria_odb10/bacteria_odb10.hmm.gz -s test/bacteria_odb10/scores_cutoff -f name -p=-1
-
reformat_pyhmmsearch -i pyhmmsearch_output.tsv -o pyhmmsearch_output.reformatted.tsv
$ pyhmmsearch -h
usage: pyhmmsearch -i <proteins.fasta> -o <output.tsv> -d
Running: pyhmmsearch v2024.4.25 via Python v3.10.14 | /Users/jolespin/miniconda3/envs/kofamscan_env/bin/python
options:
-h, --help show this help message and exit
I/O arguments:
-i PROTEINS, --proteins PROTEINS
path/to/proteins.fasta. stdin does not stream and loads everything into memory. [Default: stdin]
-o OUTPUT, --output OUTPUT
path/to/output.tsv [Default: stdout]
--no_header No header
Utility arguments:
-p N_JOBS, --n_jobs N_JOBS
Number of threads to use [Default: 1]
HMMSearch arguments:
-s SCORES_CUTOFF, --scores_cutoff SCORES_CUTOFF
path/to/scores_cutoff.tsv [id_hmm]<tab>[score_threshold], No header.
-f {accession,name}, --hmm_marker_field {accession,name}
HMM reference type (accession, name) [Default: accession]
-t SCORE_TYPE, --score_type SCORE_TYPE
{full, domain} [Default: full]
-m {gathering,noise,e,trusted}, --threshold_method {gathering,noise,e,trusted}
Cutoff threshold method [Default: e]
-e EVALUE, --evalue EVALUE
E-value threshold [Default: 10.0]
Database arguments:
-d HMM_DATABASE, --hmm_database HMM_DATABASE
path/to/database.hmm cannot be used with -b/-serialized_database
-b SERIALIZED_DATABASE, --serialized_database SERIALIZED_DATABASE
path/to/database.pkl cannot be used with -d/--database_directory. Database should be pickled dictionary {name:hmm}
Copyright 2024 Josh L. Espinoza (jolespin@newatlantis.io)
-
From pyhmmsearch:
id_protein id_hmm threshold score bias best_domain-score best_domain-bias e-value SRR13615825__k127_453760_1 PF00389.34 (24.600000381469727, 24.600000381469727) 93.686 6.702 89.856 6.702 1.984e-27 SRR13615825__k127_295655_1 PF00389.34 (24.600000381469727, 24.600000381469727) 83.195 0.005 83.167 0.005 3.456e-24 SRR13615825__k127_218710_3 PF00389.34 (24.600000381469727, 24.600000381469727) 42.235 0.004 42.073 0.004 1.559e-11 SRR13615825__k127_272080_1 PF00389.34 (24.600000381469727, 24.600000381469727) 24.673 0.000 22.067 0.000 4.154e-06 SRR13615825__k127_297426_1 PF02826.23 (25.100000381469727, 25.100000381469727) 170.426 0.003 170.122 0.003 6.392e-51 -
From reformat_pyhmmsearch:
id_protein number_of_hits ids evalues scores SRR13615825__k127_453760_1 3 ['PF00389.34', 'PF02826.23', 'PF03446.19'] [1.984e-27, 2.113e-39, 2.41e-08] [93.686, 132.902, 32.336] SRR13615825__k127_295655_1 2 ['PF00389.34', 'PF02826.23'] [3.456e-24, 7.794e-21] [83.195, 72.421] SRR13615825__k127_218710_3 1 ['PF00389.34'] [1.559e-11] [42.235] SRR13615825__k127_272080_1 2 ['PF00389.34', 'PF02826.23'] [4.154e-06, 2.035e-41] [24.673, 139.471] SRR13615825__k127_297426_1 1 ['PF02826.23'] [6.392e-51] [170.426] -
From reformat_pyhmmsearch with -b/--best_hits_only:
id_protein id evalue score SRR13615825__k127_453760_1 PF02826.23 2.113e-39 132.902 SRR13615825__k127_295655_1 PF00389.34 3.456e-24 83.195 SRR13615825__k127_218710_3 PF00389.34 1.559e-11 42.235 SRR13615825__k127_272080_1 PF02826.23 2.035e-41 139.471 SRR13615825__k127_297426_1 PF02826.23 6.392e-51 170.426
-
Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011 Oct;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. Epub 2011 Oct 20. PMID: 22039361; PMCID: PMC3197634.
-
Larralde M, Zeller G. PyHMMER: a Python library binding to HMMER for efficient sequence analysis. Bioinformatics. 2023 May 4;39(5):btad214. doi: 10.1093/bioinformatics/btad214. PMID: 37074928; PMCID: PMC10159651.
The code for PyHMMSearch is licensed under an MIT License
Please contact jolespin@newatlantis.io regarding any licensing concerns.