In this repository, you can find a re-implementation of CrossWalk: Fairness-Enhanced Node Representation Learning by Khajehnejad et al. (2022). It can be used to reproduce the results and extend on the ideas of the paper. The repository was created as part of a reproducibility study and was accepted for the ML Reproducibility Challenge 2022. For the reproducibility study itself see our Open Review Page.
Please cite the original paper if you find this useful:
@article{DBLP:journals/corr/abs-2105-02725,
author = {Ahmad Khajehnejad and
Moein Khajehnejad and
Mahmoudreza Babaei and
Krishna P. Gummadi and
Adrian Weller and
Baharan Mirzasoleiman},
title = {CrossWalk: Fairness-enhanced Node Representation Learning},
journal = {CoRR},
volume = {abs/2105.02725},
year = {2021},
url = {https://arxiv.org/abs/2105.02725},
eprinttype = {arXiv},
eprint = {2105.02725},
timestamp = {Fri, 14 May 2021 12:13:30 +0200},
biburl = {https://dblp.org/rec/journals/corr/abs-2105-02725.bib},
bibsource = {dblp computer science bibliography, https://dblp.org}
}
The following authors have contributed equally:
- Kieron Kretschmar
- Luc Sträter
- Eric Zila
- Jonathan Gerbscheid
This code was tested using the environment we provide in crosswalk.yml.
We recommend using that environment to work with the code. Additionally, we recommend installing pytorch seperately, see section below.
cd crosswalk_reproduction
conda env create -f crosswalk.yml
conda activate crosswalk
The package makes use of the pytorch framework so please first install pytorch in this new environment according to the official instructions at https://pytorch.org/get-started/locally/.
We developed the package on pytorch version 1.13.1, but newer versions are likely also supported.
The code itself is structured as a package as well. Now that we have setup our environment it is possible to install the reproduction package.
We recommend installing the package as a local package by specificying the -e . option to enable further development and access to the data. Run the following command in the root folder of the package.
pip install -e .
On Ubuntu you might need to add the path to your conda environment to your LD_LIBRARY_PATH. Inside your environment run:
export LD_LIBRARY_PATH=/path/to/anaconda3/envs/crosswalk/lib:$LD_LIBRARY_PATH
You can add this line to your .bashrc file for a permanent fix.
To reproduce the results of the original paper, we only need to the following command:
xwalk_reprod --run-all path/to/experiments/folder
where path/to/experiments/folder is the path to the folder containing the experiment config yml files included in the repository, such as experiments/node2vec
This command will iterate through all the configs in the folder and run multiple trials per experiment to calculate average and standard devation. YAML files can be grouped together in folders to do parameter sweeps, see for example experiments/param_sweeps/rice/alpha_sweep.
The experiment embeddings, graph and logs can be found in the generated results\<experiment_number> folder. The final results will also be written to a csv file in the root directory called run_all_results_<timestamp>.csv.
It is also possible to reproduce individual experiments by passing the config file as follows:
xwalk_reprod --cfg path/to/experiments/file.yml
Each experiment.yml file contains the relevant parameters for the experiment and use those to overwrite the default arguments found in config/default.py. Any parameters not present in the .yml file will be pulled form config/default.py.
You can also overwrite any of the parameters in the config or the defaults in the command line by passing the opts argument:
xwalk_reprod --cfg path/to/experiments/file.yml --opts RUNS 2 EMBEDDINGS.EMBEDDING_DIM 32
A notebook for plotting the results from a results csv file can be found in the notebooks folder. Furthermore we've included the notebook used for generation of 2d Projections and graph visualizations.
All datasets are included in this repository in the data/ folder as the datasets are reasonably sized to be included in the repository.
Pretrained embeddings can be found in data/embeddings.
While the graphs used in this repo can be read from the source dataset files in data/immutable, we have also included the dgl graphs files that can be read directly using dgl.
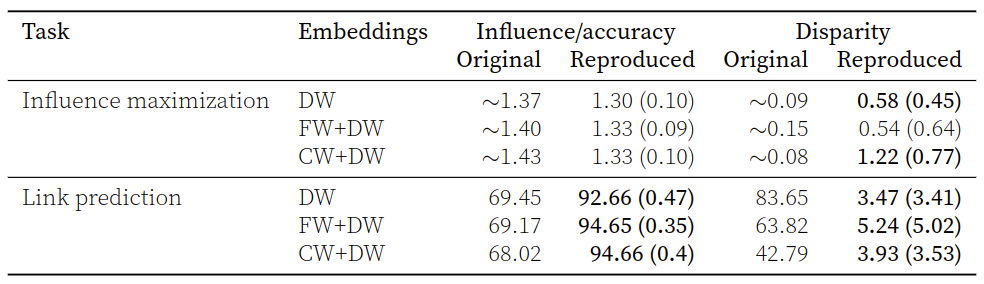
We achieve the following reproduction results:
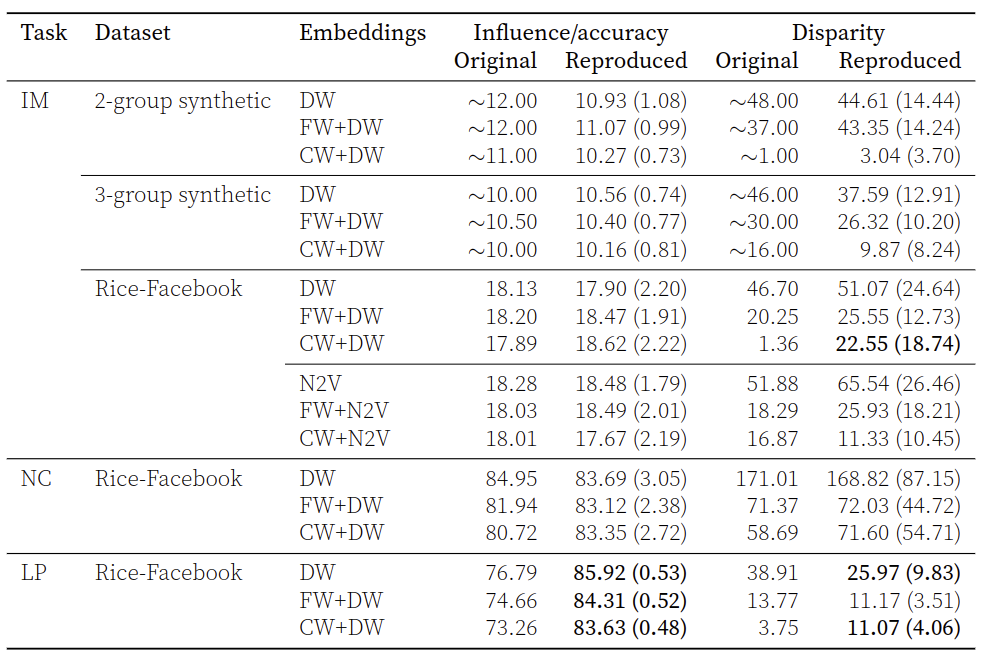
Reproduction results for twitter dataset:
To replicate the results of the original paper we make use of the gensim library. We additionally provide a pytorch implementation of node2vec that can be extended more easily.
We make use of the dgl library for our graphs. This framework is compatible with multiple popular deep learning frameworks such as Pytorch, Tensorflow & MXNet.
It is also possible to use the functions of the package by importing it. In this section we will look at a few examples of how to use the functionality provided by the package. Generally more information can be found in the docstring of each function including a description of the function, all its arguments and the output.
It is possible to read graphs in .links format and return a dgl graph.
import crosswalk_reproduction as cr
graph = cr.data_provider.read_graph("path/to/graph/graph.links")
Once we have a dgl graph loaded we can apply crosswalk or fairwalk to the weights of the graph as follows:
alpha = 0.5
p = 1
walk_length = 5
walks_per_node = 1000
group_key = 'groups' # key in graph.ndata where protected attribute is stored
prior_weights_key = 'prior_weights' # (optional) key in graph.edata where prior weights are stored
adjusted_weights = cr.preprocessor.get_crosswalk_weights(graph, alpha, p, walk_length, walks_per_node, group_key, prior_weights_key)
# or
adjusted_weights = cr.preprocessor.get_fairwalk_weights(graph, group_key)
graph.edata['adjusted_weights'] = adjusted_weights
To generate synthetic datasets, run:
import crosswalk_reproduction as cr
node_counts = [200, 300]
edge_probabilities = [[0.8, 0.8], [0.8, 0.8]]
self_connection_prob=0.0
directed=True
init_weights_strategy="uniform" # optional
weight_key="weights" # optional
remove_isolated=True # optional
group_key="groups" # optional
graph = cr.data_provider.synthesize_graph(
node_counts,
edge_probabilities,
self_connection_prob=self_connection_prob,
directed=directed,
init_weights_strategy=init_weights_strategy,
weight_key=weight_key,
remove_isolated=remove_isolated,
group_key=group_key)
For more information see the docstring of the function
To train node embeddings for a DGL graph, run:
import crosswalk_reproduction as cr
# define the needed parameters
# - for more information about these parameters check config/defaults.py
embedding_args = {
"method": "node2vec",
"embedding_dim": 32,
"walk_length": 40,
"context_size": 10,
"walks_per_node": 80,
"num_negative_samples": 1,
"p": 1.0,
"q": 1.0,
"num_workers": 4,
"lr": 0.025,
"num_epochs": 5,
"weight_key": 'weights',
"min_count": 0
}
# read graph
graph = cr.data_provider.read_graph("/path/to/graph/folder/rice_subset.links")
embeddings = cr.node_embeddor.generate_embeddings(graph, embedding_args)
# assign to graph
graph.ndata['embeddings'] = embedding.to(graph.device)
All content in the repository is licensed under the MIT license. More information can be found in the license file.
- Khajehnejad et al. (2022) - original paper
- Papers with code (2020) - README.md template