The aim of the crosscut metadata model is to provide a uniform encoding of metadata obtained from the following DCPPC data sources:
- AGR - Alliance of Genome Resources
- GTEx - Genotype-Tissue Expression project
- TOPMed - Trans-Omics for Precision Medicine
The metadata model specifies how the various metadata will be transformed into a uniform representation, whereas the metadata model instance is the transformed representation itself. The metadata model is based on a JSON-LD encoding of DATS, the DatA Tag Suite data model developed through the Big Data To Knowledge (BD2K) initiative to support dataset discoverability. See below for a description of how each of the 3 main datasets' metadata are encoded in DATS.
The crosscut metadata model instance, which is essentially a small set of JSON-LD files, is distributed as a BDBag. BDBags for all current releases can be found in the releases/ subdirectory. Each BDBag is a gzipped tar file that can be retrieved, extracted and uncompressed with standard Unix or Mac OS command line utilities. On a Mac, for example, the latest (as of this writing) v0.3 release can be retrieved and uncompressed with the following commands:
$ curl -s -O 'https://raw.githubusercontent.com/dcppc/crosscut-metadata/master/releases/KC7-crosscut-metadata-v0.3.tgz'
$ tar xzvf KC7-crosscut-metadata-v0.3.tgz
x KC7-crosscut-metadata-v0.3/
x KC7-crosscut-metadata-v0.3/tagmanifest-md5.txt
x KC7-crosscut-metadata-v0.3/bagit.txt
x KC7-crosscut-metadata-v0.3/bag-info.txt
x KC7-crosscut-metadata-v0.3/tagmanifest-sha256.txt
x KC7-crosscut-metadata-v0.3/manifest-md5.txt
x KC7-crosscut-metadata-v0.3/data/
x KC7-crosscut-metadata-v0.3/data/datasets/
x KC7-crosscut-metadata-v0.3/data/datasets/TOPMed_phs000946_wgs_public.json
x KC7-crosscut-metadata-v0.3/data/datasets/GTEx_v7_rnaseq_public.json
x KC7-crosscut-metadata-v0.3/data/datasets/MGD_GRCm38-C57BL6J_public.json
x KC7-crosscut-metadata-v0.3/data/docs/
x KC7-crosscut-metadata-v0.3/data/docs/RELEASE_NOTES
x KC7-crosscut-metadata-v0.3/data/docs/ChangeLog
x KC7-crosscut-metadata-v0.3/manifest-sha256.txt
After uncompressing the DATS JSON-LD files can be found in KC7-crosscut-metadata-v0.3/data/datasets:
$ ls -al KC7-crosscut-metadata-v0.3/data/datasets/
total 461888
drwxr-xr-x 5 jcrabtree staff 160 Jun 27 15:35 .
drwxr-xr-x 4 jcrabtree staff 128 Jun 27 15:35 ..
-rw-r--r-- 1 jcrabtree staff 48188564 Jun 27 15:35 GTEx_v7_rnaseq_public.json
-rw-r--r-- 1 jcrabtree staff 188240933 Jun 27 15:35 MGD_GRCm38-C57BL6J_public.json
-rw-r--r-- 1 jcrabtree staff 52537 Jun 27 15:35 TOPMed_phs000946_wgs_public.json
If this sounds like too much work (it isn't), the most recent raw JSON files can also be found in the dats-json/ subdirectory of this repository, in gzipped format.
The script to build the public crosscut metadata model instance is called make-crosscut-instance-bdbag.sh
and can be found in the top level of this repository:
https://github.com/dcppc/crosscut-metadata/blob/master/make-crosscut-instance-bdbag.sh
The script contains the commands to perform the DATS metadata conversion for each of the currently supported data (sub)sets, but as the comments in the file indicate, the metadata flat files for each of the data sources must first be downloaded to the current directory:
The script includes curl commands to download the requisite MGI flat files, but those commands are
commented out by default:
mkdir -p mgd-data
cd mgd-data
curl -O http://www.informatics.jax.org/downloads/mgigff/MGI.gff3.gz
curl -O http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt
cd ..
For GTEx the following two files are needed from https://www.gtexportal.org/home/datasets:
GTEx_v7_Annotations_SubjectPhenotypesDS.txt
GTEx_v7_Annotations_SampleAttributesDS.txt
For the example TOPMed study, phs000946, the public TOPMed metadata/variable summaries should be
downloaded from the following URL into a local directory named phs000946.v3:
ftp://ftp.ncbi.nlm.nih.gov/dbgap/studies/phs000946/phs000946.v3.p1/pheno_variable_summaries/
In order to run the part of the script that creates the BDBag, the bdbag command-line utility
must be installed:
pip install bdbag
The script mentioned above, make-crosscut-instance-bdbag.sh, also contains an example command showing
how to generate DATS JSON for the access-restricted metadata associated with the example TOPMed study,
phs000946. Simply add the access-restricted dbGaP files to the same local directory as the public
files (or, even better, place them in a separate directory with appropriate access controls) and then
tell the conversion script where to find the public and access-restricted metadata files, as in the
following example command:
./bin/topmed_to_dats.py --dbgap_public_xml_path=./phs000946.v3 --dbgap_protected_metadata_path=./phs000946.v3 \
--output_file=$EXTERNAL_ID/metadata/annotations/datasets/TOPMed_phs000946_wgs_RESTRICTED.json
All of the DATS JSON-LD files produced by the scripts have been validated using the validator provided
in the main DATS repository, https://github.com/datatagsuite/WG3-MetadataSpecifications. Any changes to
the DATS JSON should be checked against the validator before creating a new release of the metadata
model instance. Note that although the DATS JSON files have been validated against the JSON schemas in
the aforementioned datatagsuite repository, they will NOT validate against the current (v2.2) DATS
release from the bioCADDIE project.
This section describes how the three datasets are currently encoded in DATS and discusses some of the tradeoffs and shortcomings of the encoding. The encoding is by no means set in stone and the process of refining and improving it is still ongoing. Concomitant adjustments are also being made to the DATS model in some cases to facilitate the encoding of some aspects of the metadata.
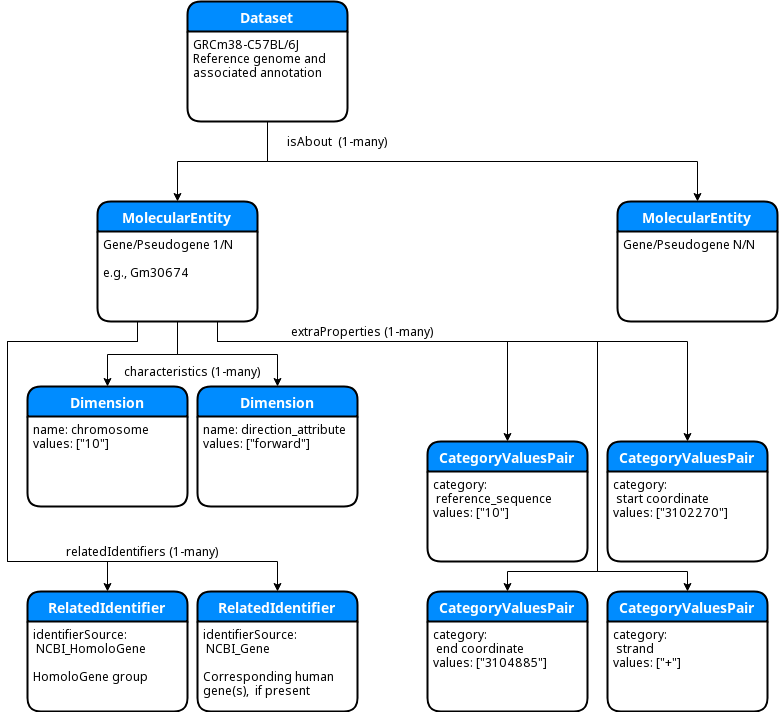
The preliminary encoding for the MGI mouse reference genome annotation is quite simple. At the top level of the
DATS JSON is a DATS Dataset object that represents the C57BL/6J reference genome and MGI gene annotation.
That top-level Dataset is linked by the isAbout property to an array of DATS MolecularEntity objects.
Each of those MolecularEntitys corresponds to an MGI gene, pseudogene, or gene/pseudogene segment. Within
each MolecularEntity the DATS properties are used as follows:
characteristics- encodes the chromosome on which the gene is located as a DATSDimensionalternateIdentifiers- lists alternate (non-MGI) ids for the gene e.g., NCBI_Gene or ENSEMBLrelatedIdentifiers- lists the HomoloGene id and any human genes linked via HomoloGeneextraProperties- contains the reference sequence id, gene coordinates, and gene strand
The following simplified ER diagram illustrates this structure:
Note that the extraProperties attribute in DATS is intended as a catch-all list for any properties that
cannot be represented in a more structured way elsewhere in the DATS entity. In general we have adopted the
position and approach that it can also be used as a place to store minimally-modified (aka "raw" or
"unharmonized") metadata from the original data source. As we improve the DATS encoding and metadata
harmonization and as the DATS model itself evolves we expect more information to appear both in "raw"
form in extraProperties and also in DATS-compliant form elsewhere in the object. A simple example of
this in the AGR/MGI encoding is the gene's reference sequence, which is encoded both in the extraProperties
(as a DATS CategoryValuesPair with category = "reference sequence") and also in the characteristics
as a DATS Dimension with a structured Annotation type drawn from a controlled vocabulary (the SO
term for "chromosome", SO_0000340.)
Notes/Comments on AGR/MGI encoding:
- The HomoloGene ids and HomoloGene-derived human gene ids in
relatedIdentifiershave DATSrelationType= http://purl.obolibrary.org/obo/SO_0000853. This is the SO term for "homologous_region". DATS only allows an IRI here, not a human-readable name and corresponding IRI, as is the case in some other places. This is perhaps not ideal because it detracts somewhat from the readability of the instance for those who don't spend their days wrangling SO terms. - DATS will soon be extended to allow a
MolecularEntityto be related to otherMolecularEntityobjects. This could be used in the MGI encoding to: 1. Directly relate genes to the chromosome on which they are found or 2. Represent human homologs as full-fledgedMolecularEntityobjects in their own right.
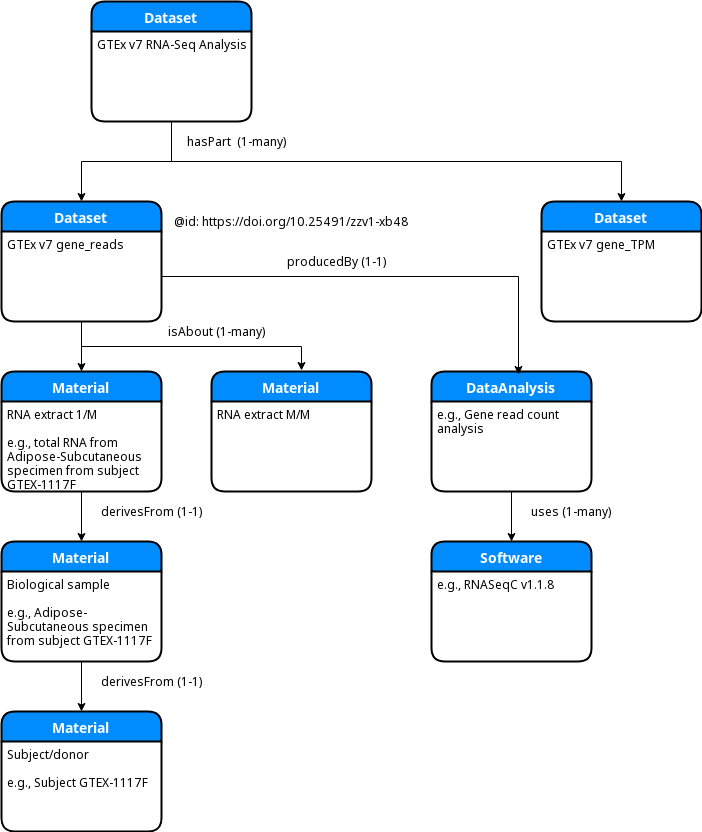
At the top level of the GTEx encoding is a DATS Dataset that represents the GTEx v7 RNA-Seq analysis.
This top level Dataset is linked by the hasPart property to an array of DATS Datasets, each of
which represents one of the public RNA-Seq data files available from https://www.gtexportal.org/home/datasets
These sub-Datasets make use of the KC2-provided DataCite GUIDs as their JSON-LD ids. For example, note
the doi.org URL in the JSON snippet below:
"@type": "Dataset",
"@context": "https://w3id.org/dats/context/sdo/dataset_context.jsonld",
"@id": "https://doi.org/10.25491/zzv1-xb48",
"identifier": {
"@type": "Identifier",
"@id": "",
"identifier": "GTEx_Analysis_2016-01-15_v7_RNA-SEQ_GTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_reads.gct.gz"
},
Each of the second level DATS Dataset objects is in turn linked to an array of DATS Material objects
by the isAbout property. Each of those Materials represents an RNA extract used in the RNA-Seq protocol.
In DATS a Material may be linked one or more other Material objects via the derivesFrom property. In
the GTEx encoding each RNA extract Material is linked first (via derivesFrom) to a Material that
represents a biological sample from a particular body site. That biological sample Material is further
linked (also via derivesFrom) to a Material that represents the individual human donor/subject, as
shown in the following ER diagram:
In the public version of the GTEx DATS encoding all of the human subjects, samples, and RNA extracts are represented, but some of the phenotype and/or sample data may be limited. For example, instead of specifying each subject's exact age, only an "Age range" (e.g,. "60-69") is provided.
Notes/Comments on GTEx encoding:
- There is a significant amount of redundancy in this encoding. Each RNA extract
Material, along with its associated biological sample and subject, is repeated for each and every one of the second-levelDatasetobjects. One alternative would be to link only the top-level RNA-SeqDatasetto the array ofMaterialsand say that the sub-Datasets are implicitly "about" thoseMaterials by dint of their relationship to the parentDataset. To our knowledge DATS itself does not require one or the other representation. Another option would be to keep the existing structure but make use of the JSON-LD ids to replace any duplicatedMaterialelements with references back to the first occurrence of each. - The gross structure of the GTEx encoding differs from that of the TOPMed encoding in the following way:
in GTEx we have a single
Datasetrepresenting the GTEx v7 RNA-Seq data with a set of sub-Datasets that represent the individual analysis products produced by analyzing the RNA-Seq data. For TOPMed, on the other hand, there is a single top-levelDatasetthat represents the umbrella TOPMed project, below which there is a second levelDatasetfor each individual study within TOPMed. There are no third levelDatasetobjects that represent the datasets produced within each study. Therefore in a future release it is likely that both the GTEx and TOPMed encodings may standardize on a 3-levelDatasetstructure at the top.
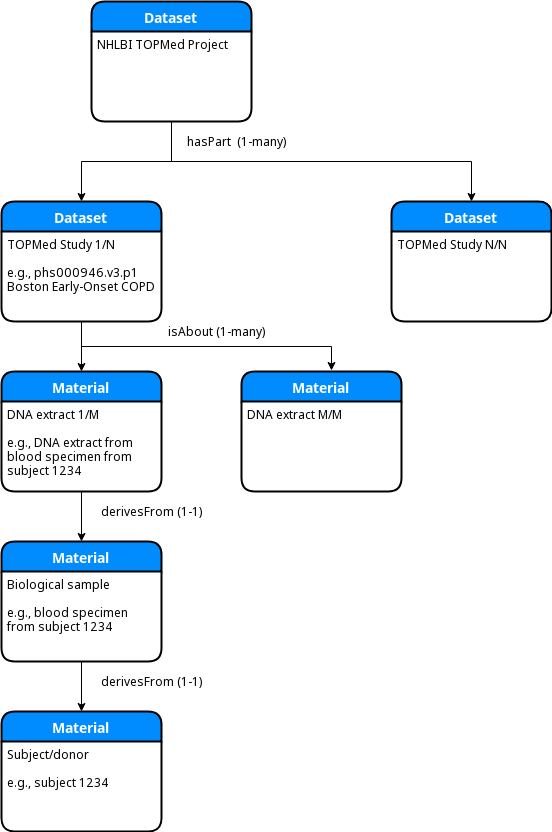
At the top level of the TOPMed encoding is an umbrella DATS Dataset that represents the overarching
TOPMed project. That Dataset links to one or more sub-Datasets via the hasPart property. These
second level Datasets represent the individual studies that comprise TOPMed. In the current instance
only one example TOPMed study is present at the second level, namely phs000946, the "Boston Early-Onset
COPD Study in the TOPMed Program" Within those second level Datasets the organization is similar to
that used in GTEx, with each Dataset linked to an array of DATS Material objects by the isAbout
property. Each of those Materials represents a DNA extract and is linked (via derivesFrom) first
to a biological sample and then (again via derivesFrom) to the human subject/donor. The following
simplified ER diagram illustrates this structure:
Notes/Comments on TOPMed encoding:
- See the GTEx notes for a description of the inconsistency between the two levels of
Datasetused at the top of the GTEx encoding versus the two levels used at the top of the TOPMed encoding.