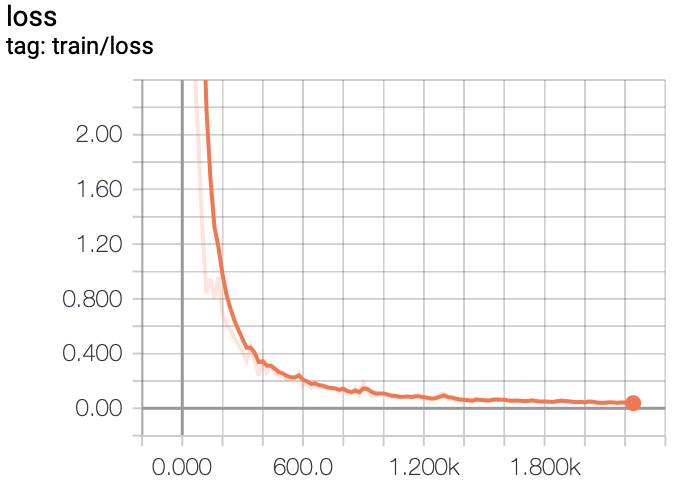
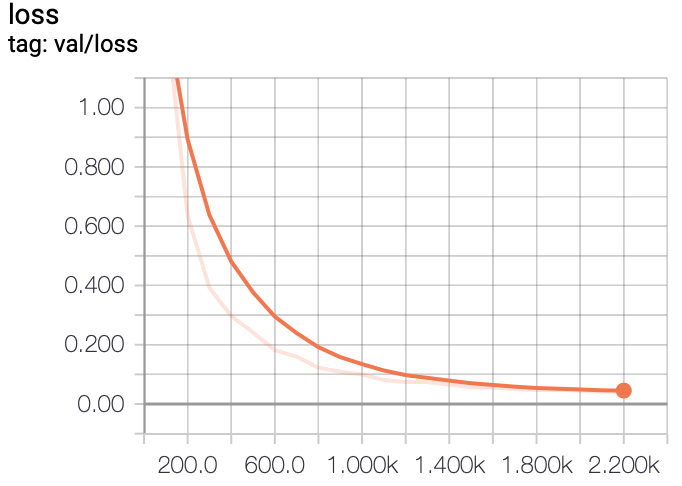
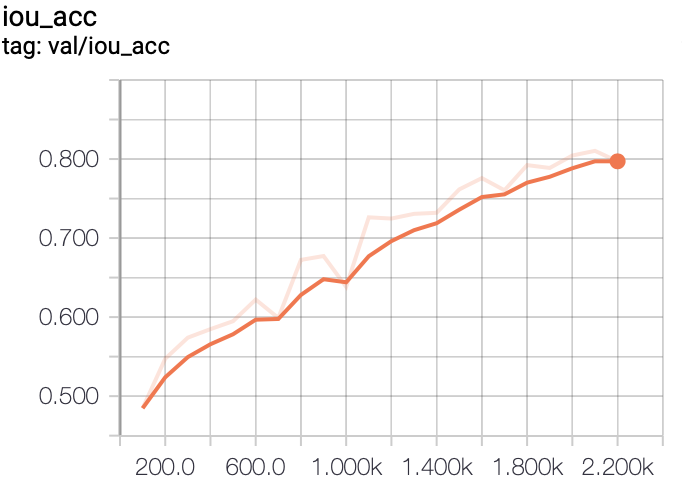
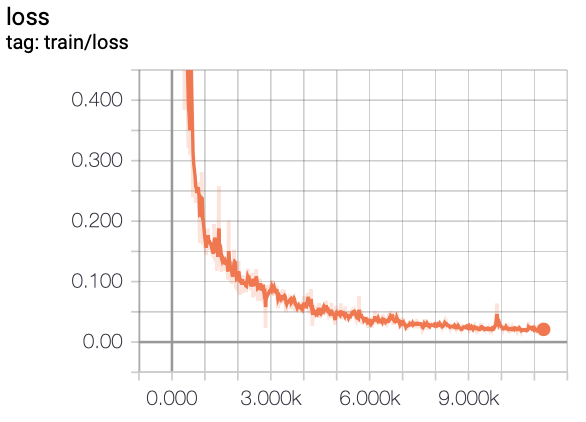
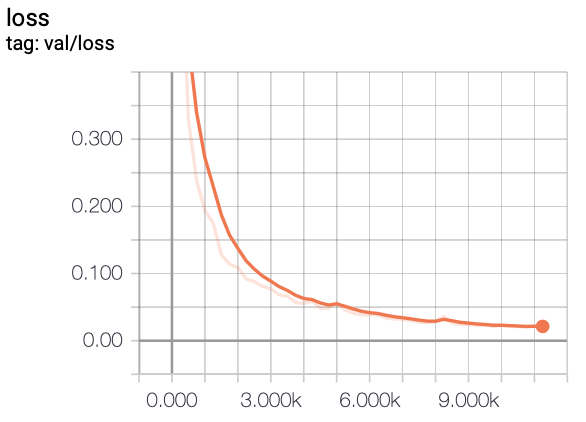
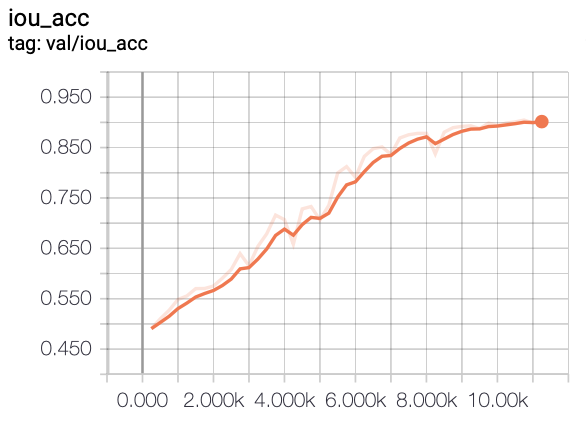
In this project, we experiment with training FCN models on a liver segmentation dataset provided by IRCAD
First, we need to set up a virtual Python 3.6 environment as follows
pip install virtualenv
virtualenv -p python3 .env
source .env/bin/activate
pip install -r requirements.txtThen, download the dataset 3D-ircadb-01 by running the script below
sh scripts/download.shThe image format in the dataset is in DICOM format so we have to do some preprocessing stuff before converting data to TFRecord format for later training in TensorFlow.
In the scope of this project, we only segment 4 classes including liver, bone, kidney and others. Therefore, each preprocessed data sample will be a pair of
- an
image- an input image of shape (512, 512) and - a
mask- a 2D array has the same spatial shape as theimageindicating which class each pixel belongs to.
python run.py --mode preprocessWe use TensorFlow iterator to iterate over the dataset. To check image and mask shape, run the command below
python run.py --mode iterTo train FCN models with default config, run
python run.py --mode trainTo predict output on the validation set, run
python run.py --mode predict- Preprocessing
- Building model
- Training pipeline
- Augmentation
- Visualization
- TensorBoard
References
- Long et al. , Fully Convolutional Networks for Semantic Segmentation. (arxiv)