Anc2vec is a novel method based on neural networks to construct embeddings of terms from the Gene Ontology (GO) exclusively using three structural features of it: the ontological uniqueness of terms, their ancestor relationships and the sub-ontology to which they belong.
This repository offers a Python package containing the source code of anc2vec, as well as instructions for reproducibility of the main results of the study where this method was proposed:
Anc2vec: embedding Gene Ontology terms by preserving ancestors relationships, by A. A. Edera, D. H. Milone, and G. Stegmayer. Research Institute for Signals, Systems and Computational Intelligence, sinc(i).
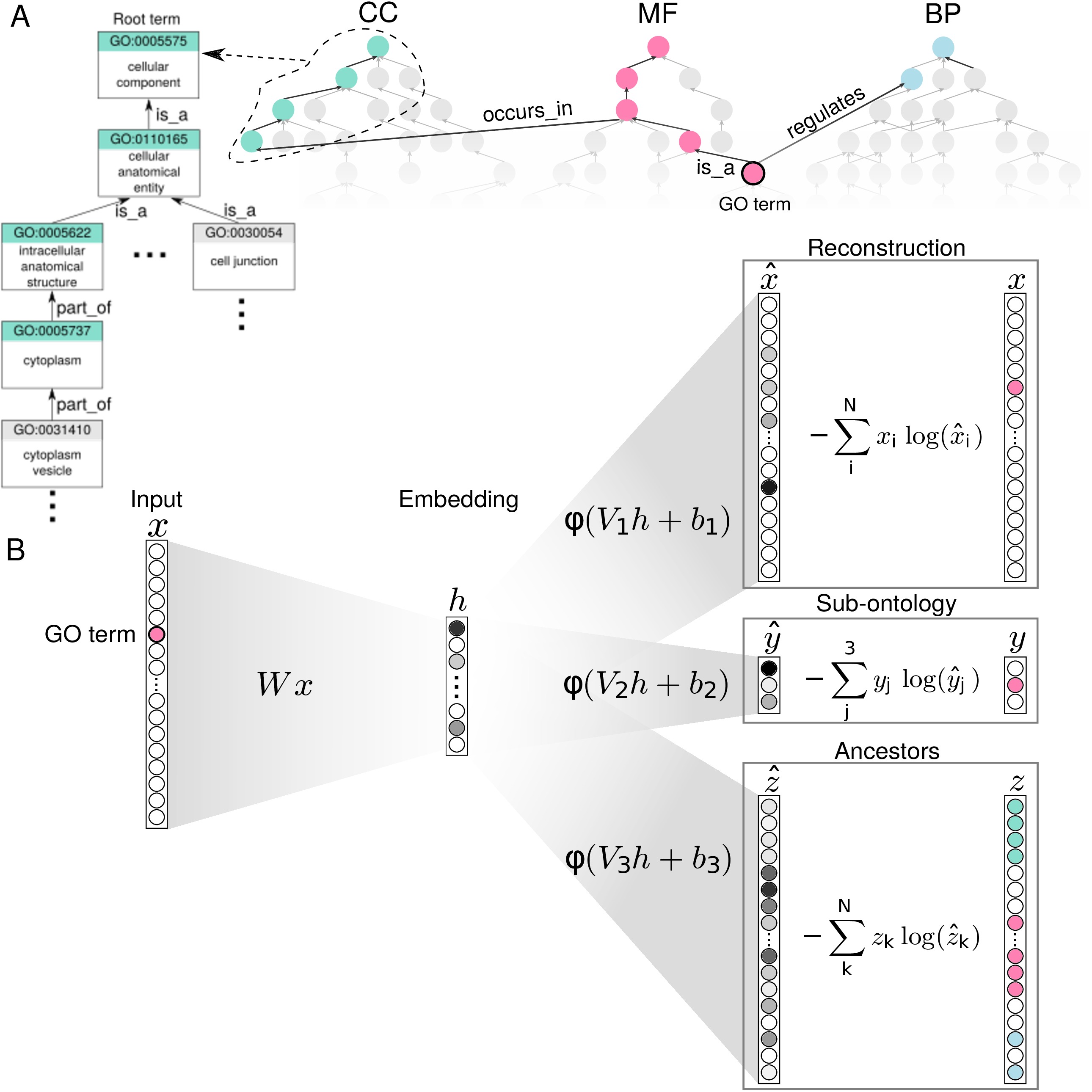
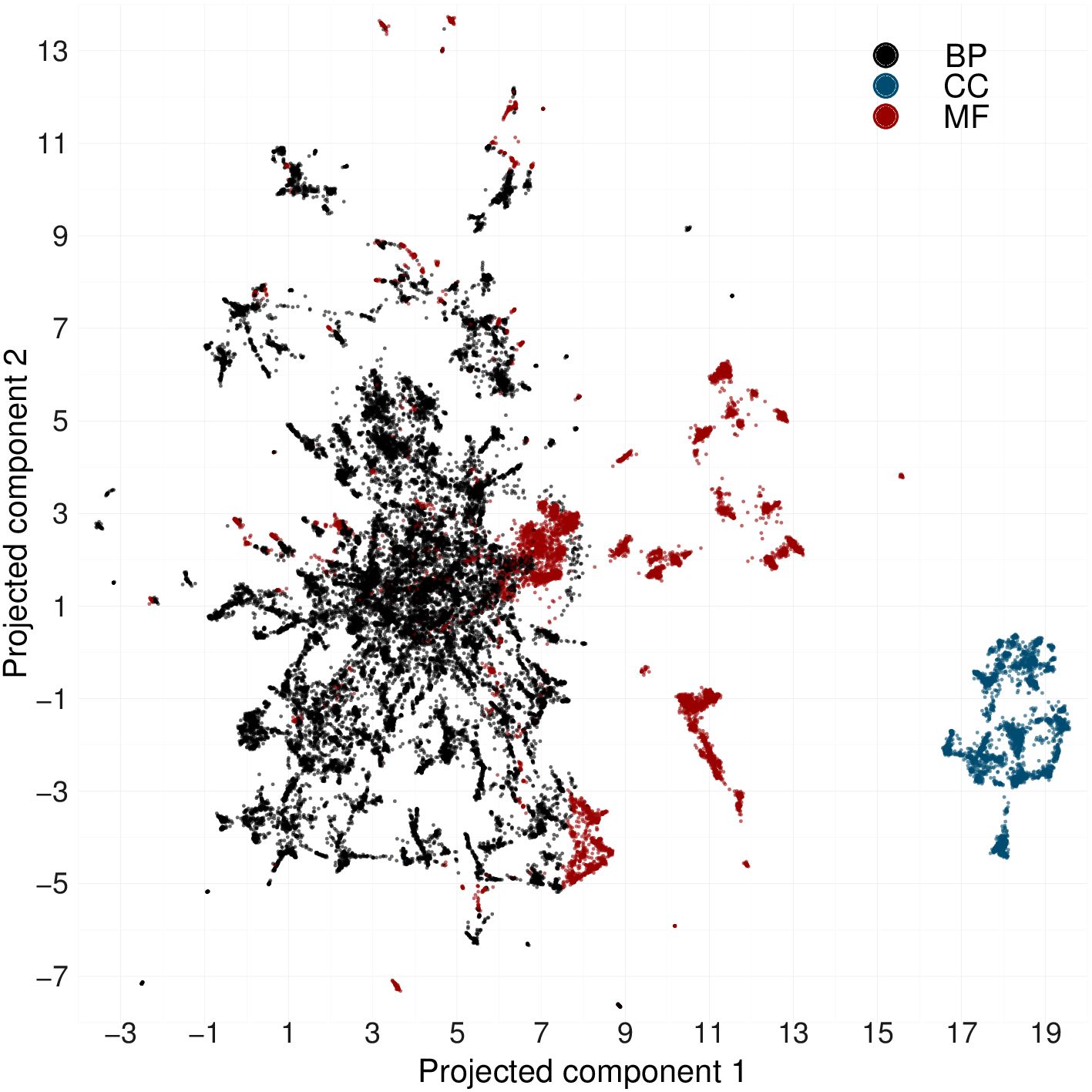
Fig. 1. Panel A) The GO structure is composed by hierarchical relationships between terms arranged in three sub-ontologies: BP, CC, and MF. Panel B) Anc2vec architecture. A GO term is encoded as a one-hot vector x that is transformed into an embedding h, which is used to predict three structural features of the GO that are used for weight optimization. Fig. 2. Anc2vec embeddings of GO terms in the three sub-ontologies. Points depict embeddings of GO terms whose colors encode the sub-ontologies: BP (Biological Process), CC (Cellular Component), and MF (Molecular Function). There is available a video showing how 2-dimensional embeddings are adjusted during weight optimization.Anc2vec requires Python 3.6 and TensorFlow 2.3.1.
It is recommendable to have installed Conda, to avoid Python package conflicts.
If Conda is installed, first create and activate a conda environment, for example, named anc2vec:
conda create --name anc2vec python=3.6
conda activate anc2vecNext, install the anc2vec package via the
pip package manager:
pip install -U "anc2vec @ git+https://github.com/aedera/anc2vec.git"The anc2vec package has already available the same embedding of GO terms
used in the study. These embeddings were built using the Gene Ontology release
2020-10-06. The embeddings can be easily accessed on
Python with this command:
import anc2vec
es = anc2vec.get_embeddings()Here, es is a python dictionary that maps GO terms with their corresponding
200-dimensional embeddings. For example, this command uses this dictionary to
retrieve the embedding corresponding to the term GO:0001780:
e = es['GO:0001780']The variable e is a Numpy array containing the
embedding
array([ 0.55203265, -0.23133564, 0.1983797 , -0.3251996 , 0.20564775,
-0.32133245, -0.25364587, -0.16675541, -0.46832997, -0.40702957,
...
-0.29757708, -0.33143485, -0.31099185, 0.24465033, -0.25458524,
-0.24525951, -0.366758 , -0.04628978, 0.29378492, 0.31249675],
dtype=float32)These anc2vec embeddings are ready to be used for semantic similarity
tasks. Below there are examples showing how to use them for calculating
cosine distances.
The anc2vec package also contains a function to build embeddings from
scratch using a specific
OBO file, a
human-readable file usually used to describe the GO. Building embeddings can
be particularly useful for experimental scenarios where a specific version of
the GO is required, such as those available in the
GO data archive.
The following code shows how to build the embedding for a given OBO file named
go.obo.
import anc2vec
import anc2vec.train as builder
es = builder.fit('go.obo', embedding_sz=200, batch_sz=64, num_epochs=100)The object builder uses the input go.obo file to extract structural
features used to build the embeddings of GO terms. Note that builder is
called with additional parameters indicating the dimensionality of the
embeddings (embedding_sz) and the number of optimization steps used for
embedding building (num_epochs). The embeddings built by builder are
stored in es, which is a Python dictionary mapping GO terms to their
corresponding embeddings.
Please check the examples below for more information about this functionality.
To try anc2vec, below there are links to Jupyter notebooks that use Google Colab which offers free computing on the Google cloud.
These are the main datasets used in the experiments of the study where anc2vec is proposed:
The anc2vec package is released under the MIT License.