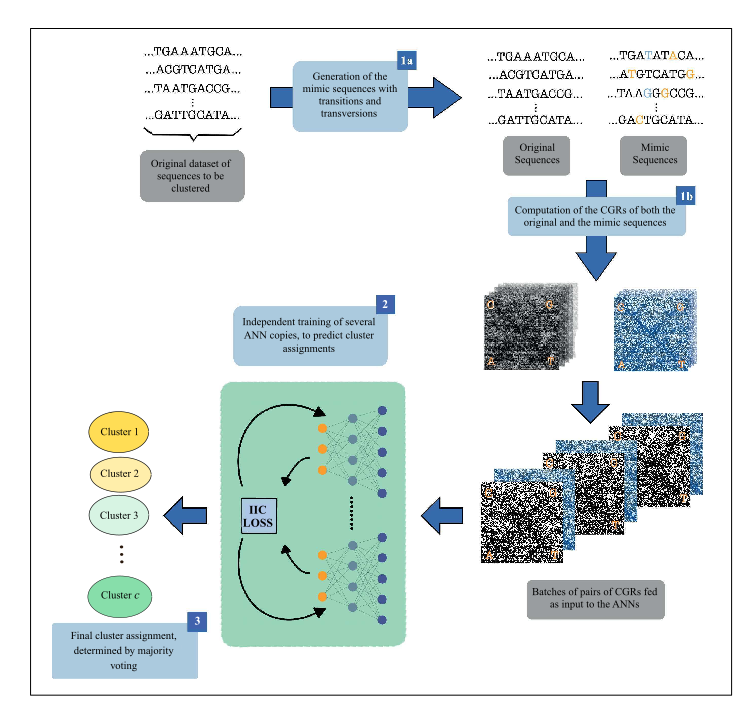
This repository contains all the source files required to run DeLUCS (https://doi.org/10.1101/2021.05.13.444008) a deep learning clustering tool for DNA sequences, as well as a detailed guide for running the code.
python build_dp.py --data_path=<PATH_sequence_folder>
- Input: Folders with the sequences in FASTA format
- Output : file in the form (label,sequence,accession)
python get_pairs.py --data_path=<PATH_pickle_dataset> --k=6 --modify='mutation' --output=<PATH_output_file> --n_mimics=<n mimics per sequence>
- Input: file in the form (label,sequence,accession)
- Output : file in the form of (pairs, x_test, y_test)
-
For training DeLUCS to cluster your own data (No ground truth available):
python TrainDeLUCS.py --n_clusters=<number of clusters> --data_dir=<PATH_of_computed_mimics> --out_dir=<OUTPURDIR>- Input: Pickle file with the mimics (Always ending with /testing_data.p) .
- Output : Pickle file with the cluster assignments for each sequence.
-
For training DeLUCS and testing its performance with your own data (labels must be available)
python EvaluateDeLUCS.py --data_dir=<PATH_of_computed_mimics> --out_dir=<OUTPURDIR>- Input: Pickle file with the mimics in the form of (pairs, x_test, y_test).
- Output : Confusion Matrix.
-
For training a single Neural Network in an unsupervised way:
python SingleRun.py --n_clusters=<number of clusters> --data_dir=<PATH_of_computed_mimics> --out_dir=<OUTPURDIR> -
For testing the performance a single Neural Network trained in an unsupervised way (labels must be available):
python EvaluateSingleRun.py --data_dir=<PATH_of_computed_mimics> --out_dir=<OUTPURDIR>