partycls is a Python package for cluster analysis of systems of interacting particles. By grouping particles that share similar structural or dynamical properties, partycls enables rapid and unsupervised exploration of the system's relevant features. It provides descriptors suitable for applications in condensed matter physics, such as structural analysis of disordered or partially ordered materials, and integrates the necessary tools of unsupervised learning into a streamlined workflow.
For more details and tutorials, visit the homepage at: https://www.jorisparet.com/partycls
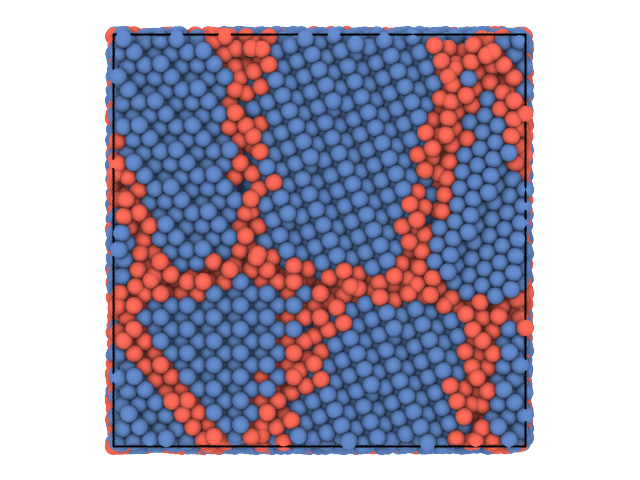
This quick example shows how to use partycls to identify grain boundaries in a polycrystalline system. The system configuration is stored in a XYZ trajectory file with a single frame. We use the local distribution of bond angles around each particle as a structural descriptor and perform a clustering using the K-Means algorithm.
from partycls import Trajectory, Workflow
traj = Trajectory('grains.xyz')
wf = Workflow(traj, descriptor='ba', clustering='kmeans')
wf.run()
traj[0].show(color='label', backend='ovito')The results are also written to a set of files including a labeled trajectory file and additional information on the clustering results. The whole workflow can be tuned and customized, check out the tutorials to see how and for further examples.
Thanks to a flexible system of filters, partycls makes it easy to restrict the analysis to a given subset of particles based on arbitrary particle properties. Say we have a binary mixture composed of particles with types A and B, and we are only interested in analyzing the bond angles of B particles in a vertical slice:
from partycls import Trajectory
from partycls.descriptors import BondAngleDescriptor
traj = Trajectory('trajectory.xyz')
D = BondAngleDescriptor(traj)
D.add_filter("species == 'B'")
D.add_filter("x > 0.0")
D.add_filter("x < 1.0")
D.compute()
# Angular correlations for the selected particles
print(D.features)We can then perform a clustering based on these structural features and ask for 3 clusters:
from partycls import KMeans
clustering = KMeans(n_clusters=3)
clustering.fit(D.features)
print('Cluster membership of the particles', clustering.labels)partycls accepts several trajectory formats (including custom ones) either through its built-in trajectory reader or via third-party packages, such as MDTraj and atooms. The code is currently optimized for small and medium system sizes (of order 10⁴ particles). Multiple trajectory frames can be analyzed to extend the structural dataset.
partycls implements various structural descriptors:
- Radial descriptor
- Tetrahedral descriptor
- Bond-angle descriptor
- Smoothed bond-angle descriptor
- Bond-orientational descriptor
- Smoothed bond-orientational descriptor
- Locally averaged bond-orientational descriptor
- Radial bond-orientational descriptor
- Compactness descriptor
- Coordination descriptor
partycls performs feature scaling, dimensionality reduction and cluster analysis using the scikit-learn package and additional built-in algorithms.
partycls relies on several external packages, most of which only provide additional features and are not necessarily required.
- Fortran compiler (e.g. gfortran)
- NumPy
- scikit-learn
- MDTraj (additional trajectory formats)
- atooms (additional trajectory formats)
- DScribe (additional descriptors)
- Matplotlib (visualization)
- OVITO < 3.7.0 (visualization)
- Py3DMol (interactive 3D visualization)
- pyvoro or its memory-optimized fork for large systems (Voronoi neighbors and tessellation)
- tqdm (progress bars)
Check the tutorials to see various examples and detailed instructions on how to run the code, as well as an in-depth presentation of the built-in structural descriptors.
For a more detailed documentation, you can check the API.
The latest stable release is available on PyPI. Install it with pip:
pip install partyclsTo install the latest development version from source, clone the source code from the official GitHub repository and install it with:
git clone https://github.com/jorisparet/partycls.git
cd partycls
make installRun the tests using:
make testor manually compile the Fortran sources and run the tests:
cd partycls/
f2py -c -m neighbors_wrap neighbors.f90
cd descriptor/
f2py -c -m realspace_wrap realspace.f90
cd ../../
pytest tests/If you wish to contribute or report an issue, feel free to contact us or to use the issue tracker and pull requests from the code repository.
We largely follow the GitHub flow to integrate community contributions. In essence:
- Fork the repository.
- Create a feature branch from
master. - Unleash your creativity.
- Run the tests.
- Open a pull request.
We also welcome contributions from other platforms, such as GitLab instances. Just let us know where to find your feature branch.
If you use partycls in a scientific publication, please consider citing the following article:
partycls: A Python package for structural clustering. Paret et al., (2021). Journal of Open Source Software, 6(67), 3723
Bibtex entry:
@article{Paret2021,
doi = {10.21105/joss.03723},
url = {https://doi.org/10.21105/joss.03723},
year = {2021},
publisher = {The Open Journal},
volume = {6},
number = {67},
pages = {3723},
author = {Joris Paret and Daniele Coslovich},
title = {partycls: A Python package for structural clustering},
journal = {Journal of Open Source Software}
}