The code for the ConGen model described in the arXiv paper "Multi-Constraint Molecular Generative Model using Sparsely Labelled Training Data: Localized High-Concentration Electrolyte Diluent Screening Use Case" by J.P. Mailoa, et al.
The full paper is published here: https://doi.org/10.1039/D3DD00064H.
This code is based on the original implementation of semi-supervised variational auto-encoder (SSVAE) model described in the paper https://pubs.acs.org/doi/abs/10.1021/acs.jcim.8b00263 by S. Kang, et al.
The main modifications implemented in this repository consist of the following:
- PyTorch implementation instead of the original TensorFlow 1.0 implementation
- Ability to train generative model using a combination of 'dirty data' with irregular missing labels
- Ability to replace individual sub-model with external pre-trained model for transfer learning, if desired
In the original implementation of the SSVAE model, the training data needs to be 'ideal'. This means the molecules in the database are either fully labeled or fully unlabeled. Minibatches of molecules will be supplied to the model during training, but this has to be done in such a way that the fully labeled molecules go through the model differently than the fully unlabeled molecules.
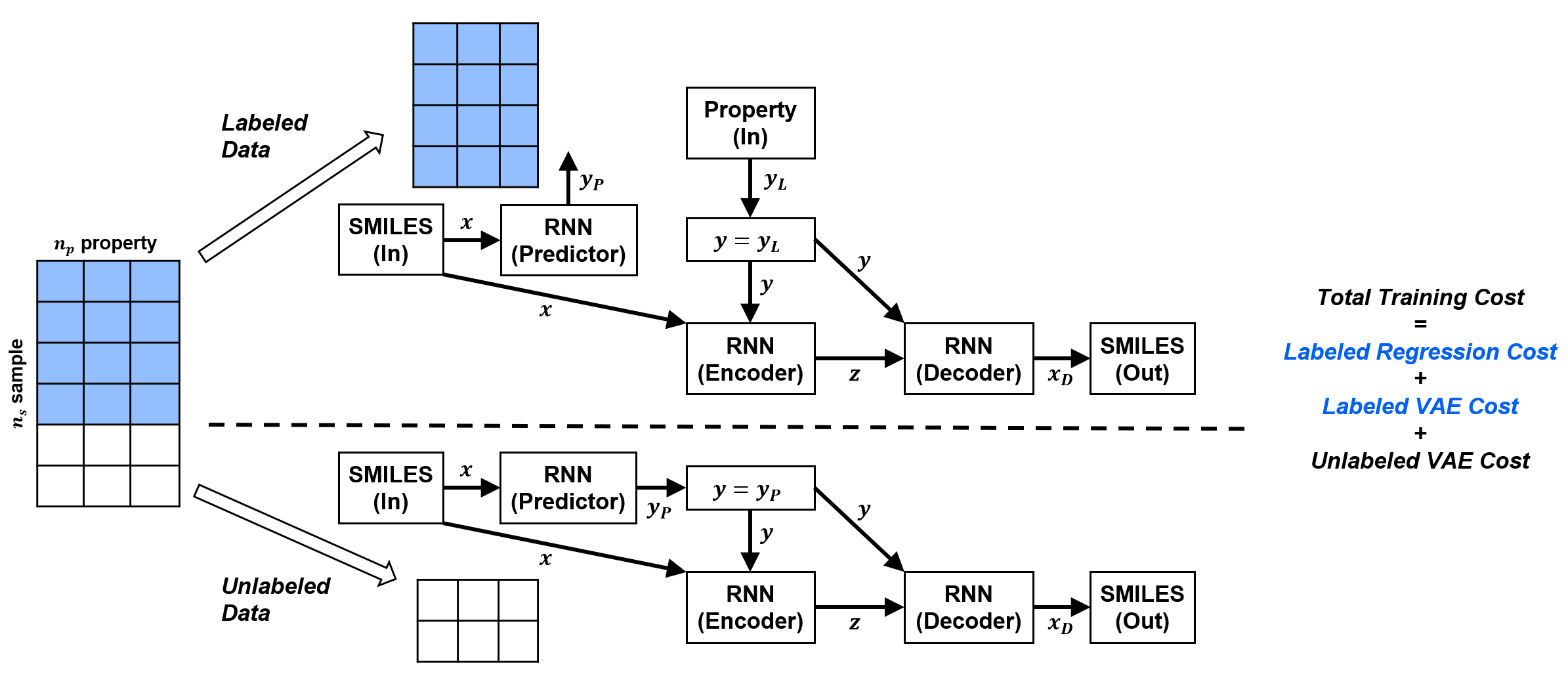
In reality, molecule label training data is not so 'clean'. When we obtain molecule-property database from publicly available sources, property labels are often patially incomplete (especially so for experimental data). Or in other cases, we have molecule property dataset from different sources, and the combined database will be partially unlabeled.
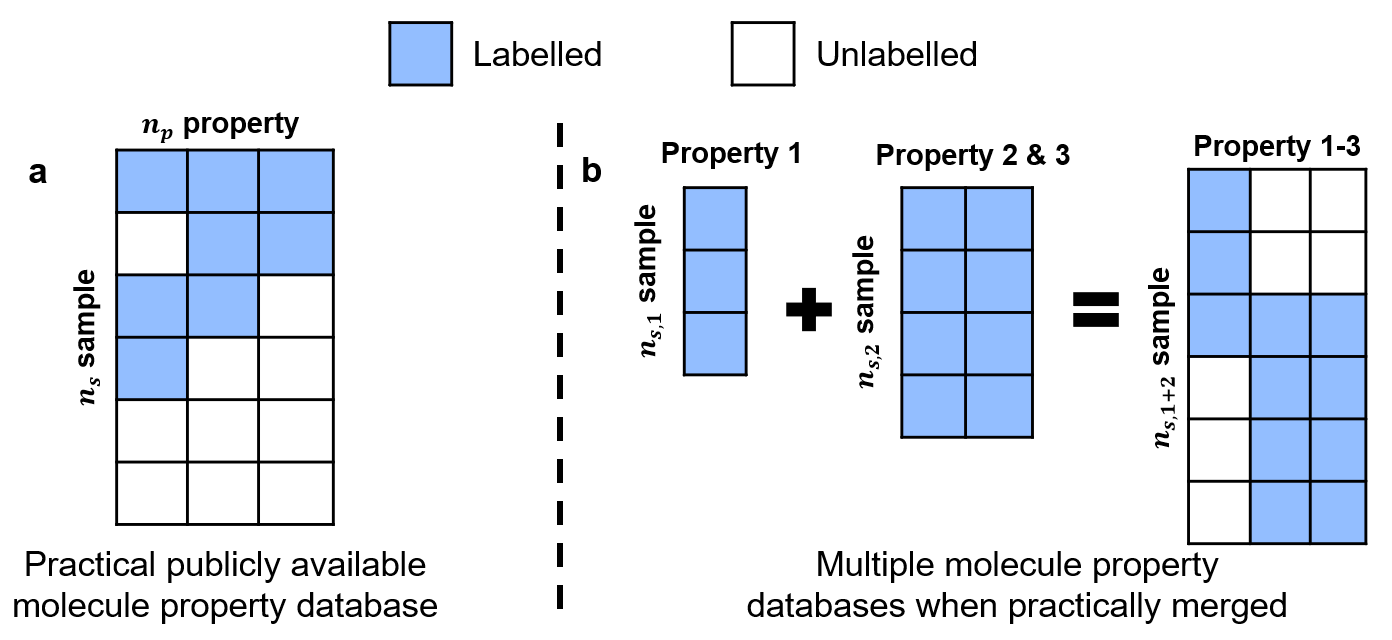
In order to remedy this, we develop a new mechanism to represent the data with incomplete label, using a mask.
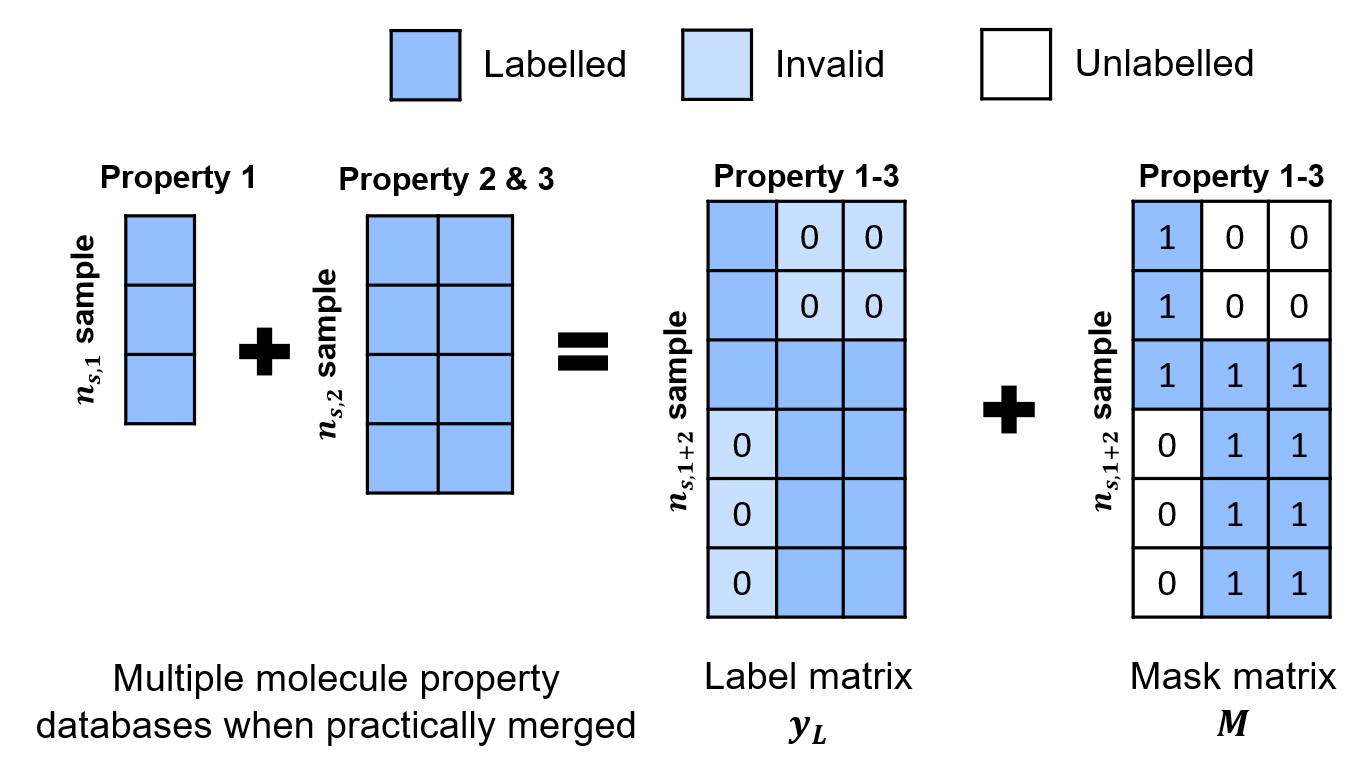
By utilizing the property mask and modifying the SSVAE model accordingly to take into account these masks, we are able to utilize all the available property labels for multi-property molecule generation tasks.
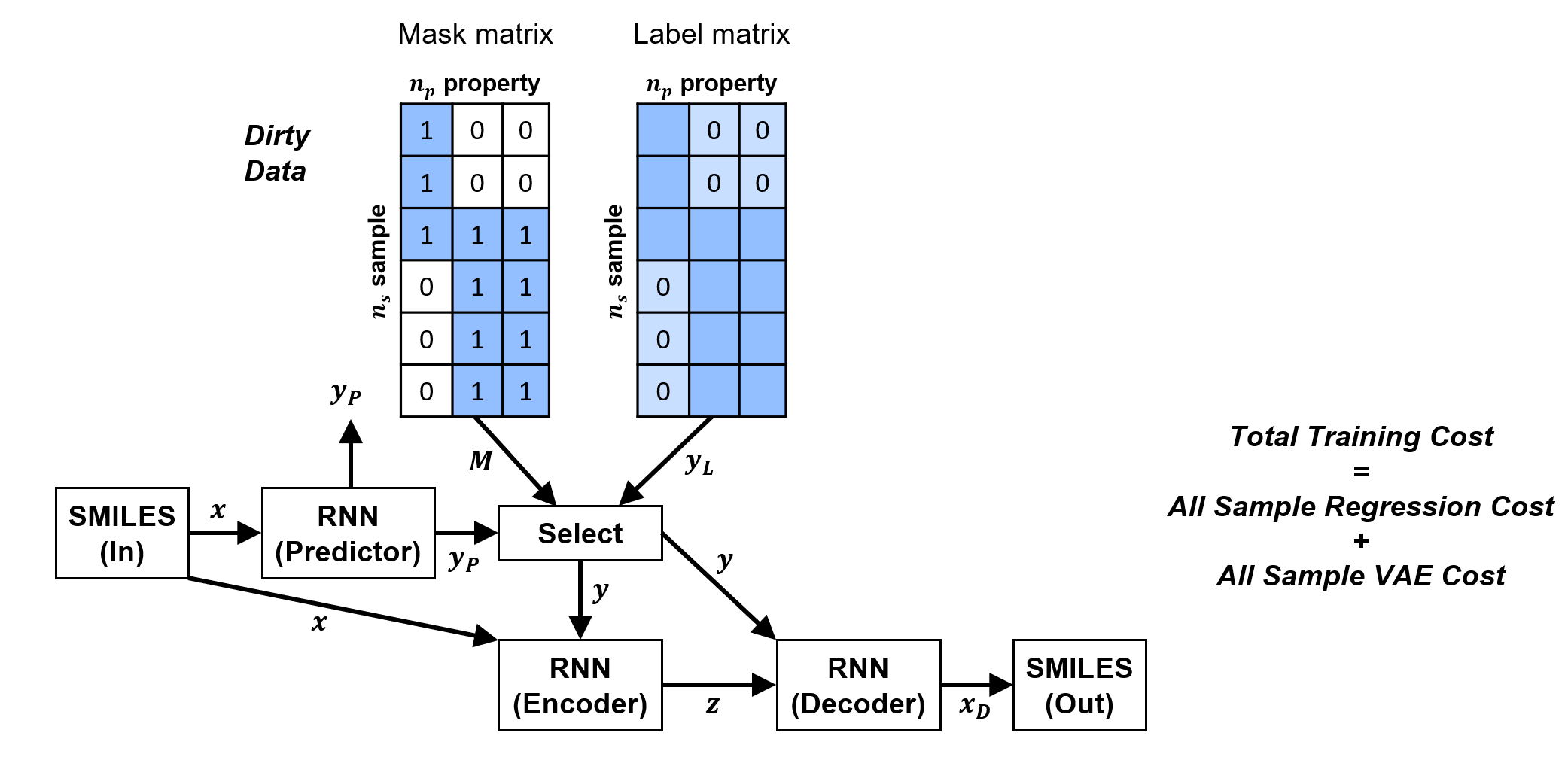
There are multiple model modifications and associated cost functions which need to be done to enable ConGen to work correctly. The details can be found within SSVAE.py and within the ConGen arXiv paper cited above.
There are two main notebooks which may reflect the typical usage of this model. train_generator.ipynb shows example on how to train the ConGen model, while repeat_generation.ipynb shows how to reload the trained model and repeatedly use it for different regression or unconditional / conditional generation purposes. To run them, respectively use:
jupyter nbconvert --to=notebook --inplace --ExecutePreprocessor.enabled=True train_generator.ipynb
jupyter nbconvert --to=notebook --inplace --ExecutePreprocessor.enabled=True repeat_generation.ipynb
Run the following instructions to conveniently generate the conda environment with the right dependencies:
conda create --name ConGen python=3.7
conda activate ConGen
conda install --file requirements.txt -c pytorch -c conda-forge -c rdkit