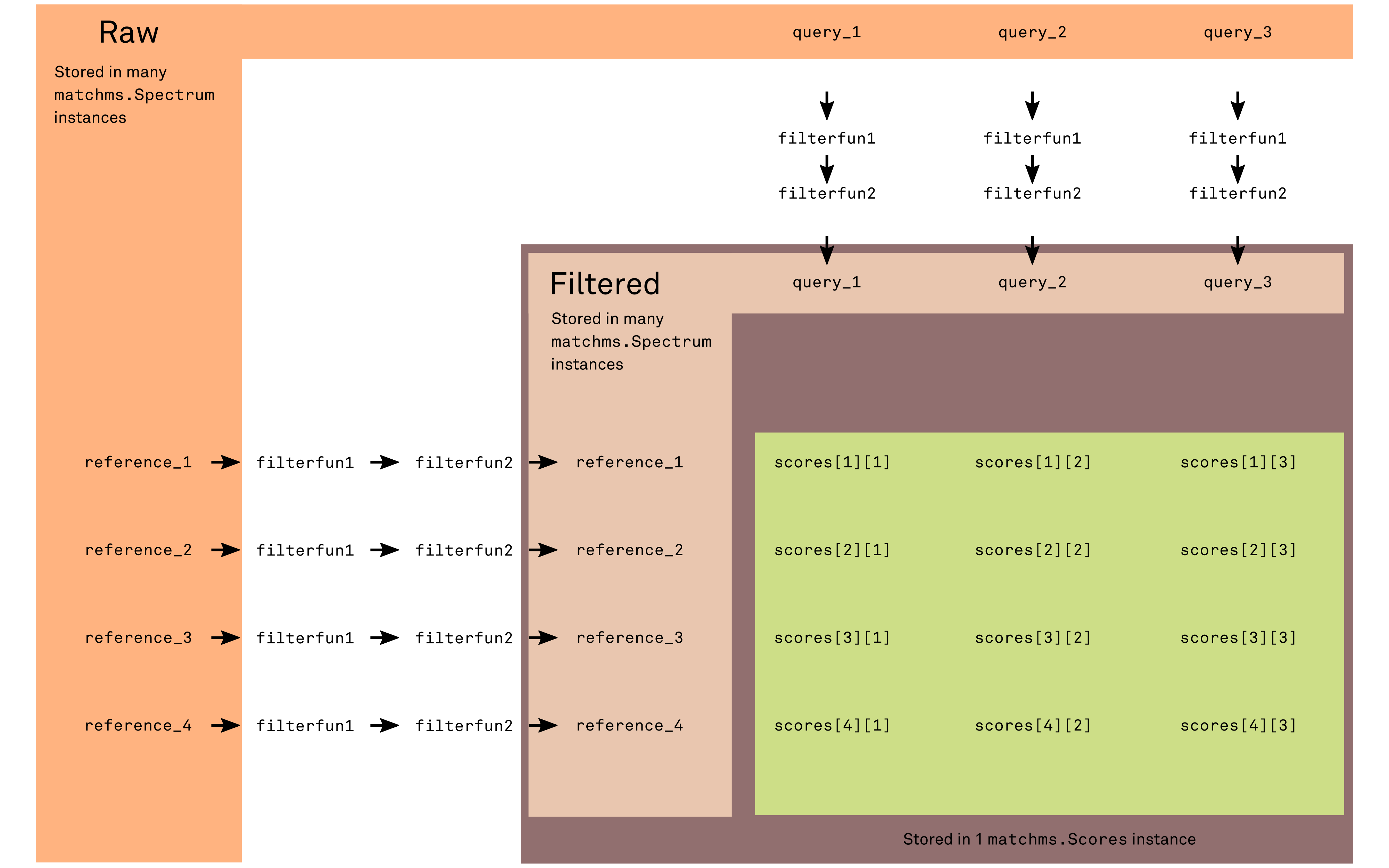
Vector representation and similarity measure for mass spectrometry data.
| fair-software.nl recommendations | Badges |
|---|---|
| 1. Code repository |  |
| 2. License |  |
| 3. Community Registry | 
|
| 4. Enable Citation | |
| 5. Checklist | |
| Other best practices | |
| Continuous integration | |
| Documentation | |
| Code Quality |
Install matchms from Anaconda Cloud with
conda install --channel nlesc --channel bioconda --channel conda-forge matchmsBelow is a small example of using matchms to calculate the Cosine score between mass Spectrums in the tests/pesticides.mgf file.
from matchms.importing import load_from_mgf
from matchms.filtering import default_filters
from matchms.filtering import normalize_intensities
from matchms import calculate_scores
from matchms.similarity import CosineGreedy
# Read spectrums from a MGF formatted file, for other formats see https://matchms.readthedocs.io/en/latest/api/matchms.importing.html
file = load_from_mgf("tests/pesticides.mgf")
# Apply filters to clean and enhance each spectrum
spectrums = []
for spectrum in file:
# Apply default filter to standardize ion mode, correct charge and more.
# Default filter is fully explained at https://matchms.readthedocs.io/en/latest/api/matchms.filtering.html .
spectrum = default_filters(spectrum)
# Scale peak intensities to maximum of 1
spectrum = normalize_intensities(spectrum)
spectrums.append(spectrum)
# Calculate Cosine similarity scores between all spectrums
# For other similarity score methods see https://matchms.readthedocs.io/en/latest/api/matchms.similarity.html .
scores = calculate_scores(references=spectrums,
queries=spectrums,
similarity_function=CosineGreedy())
# Print the calculated scores for each spectrum pair
for score in scores:
(reference, query, score, n_matching) = score
# Ignore scores between same spectrum and
# pairs which have less than 20 peaks in common
if reference != query and n_matching >= 20:
print(f"Reference scan id: {reference.metadata['scans']}")
print(f"Query scan id: {query.metadata['scans']}")
print(f"Score: {score:.4f}")
print(f"Number of matching peaks {n_matching}")
print("----------------------------")| Term | Description |
|---|---|
| adduct / addition product | During ionization in a mass spectrometer, the molecules of the injected compound break apart into fragments. When fragments combine into a new compound, this is known as an addition product, or adduct. Wikipedia |
| GNPS | Knowledge base for sharing of mass spectrometry data (link). |
InChI / INCHI |
InChI is short for International Chemical Identifier. InChIs are useful in retrieving information associated with a certain molecule from a database. |
InChIKey / InChI key / INCHIKEY |
An indentifier for molecules. For example, the InChI key for carbon
dioxide is InChIKey=CURLTUGMZLYLDI-UHFFFAOYSA-N (yes, it
includes the substring InChIKey=). |
| MGF File / Mascot Generic Format | A plan ASCII file format to store peak list data from a mass spectrometry experiment. Links: matrixscience.com, fiehnlab.ucdavis.edu. |
parent mass / parent_mass |
Actual mass (in Dalton) of the original compound prior to fragmentation. It can be recalculated from the precursor m/z by taking into account the charge state and proton/electron masses. |
precursor m/z / precursor_mz |
Mass-to-charge ratio of the compound targeted for fragmentation. |
| SMILES | A line notation for describing the structure of chemical species using
short ASCII strings. For example, water is encoded as O[H]O,
carbon dioxide is encoded as O=C=O, etc. SMILES-encoded species may be converted to InChIKey using a resolver like this one. The Wikipedia entry for SMILES is here. |
To install matchms, do:
git clone https://github.com/matchms/matchms.git
cd matchms
conda env create --file conda/environment-dev.yml
conda activate matchms-dev
pip install --editable .Run the linter with:
prospectorAutomatically fix incorrectly sorted imports:
isort --recursive .Files will be changed in place and need to be committed manually.
Run tests (including coverage) with:
pytestTo build anaconda package locally, do:
conda deactivate
conda env create --file conda/environment-build.yml
conda activate matchms-build
BUILD_FOLDER=/tmp/matchms/_build
rm -rfv $BUILD_FOLDER;mkdir -p $BUILD_FOLDER
conda build --numpy 1.18.1 --no-include-recipe -c bioconda -c conda-forge \
--croot $BUILD_FOLDER ./condaIf successful, this will yield the built matchms conda package as
matchms-<version>*.tar.bz2 in $BUILD_FOLDER/noarch/. You can test if
installation of this conda package works with:
# make a clean environment
conda deactivate
cd $(mktemp -d)
conda env create --name test python=3.7
conda activate test
conda install \
--channel bioconda \
--channel conda-forge \
--channel file://${CONDA_PREFIX}/noarch/ \
matchmsTo publish the package on anaconda cloud, do:
anaconda --token ${{ secrets.ANACONDA_TOKEN }} upload --user nlesc --force $BUILD_FOLDER/noarch/*.tar.bz2where secrets.ANACONDA_TOKEN is a token to be generated on the Anaconda Cloud website. This secret should be added to GitHub repository.
To remove matchms package from the active environment:
conda remove matchmsTo remove matchms-build environment:
conda env remove --name matchms-buildIf you want to contribute to the development of matchms, have a look at the contribution guidelines.
Copyright (c) 2020, Netherlands eScience Center
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
This package was created with Cookiecutter and the NLeSC/python-template.