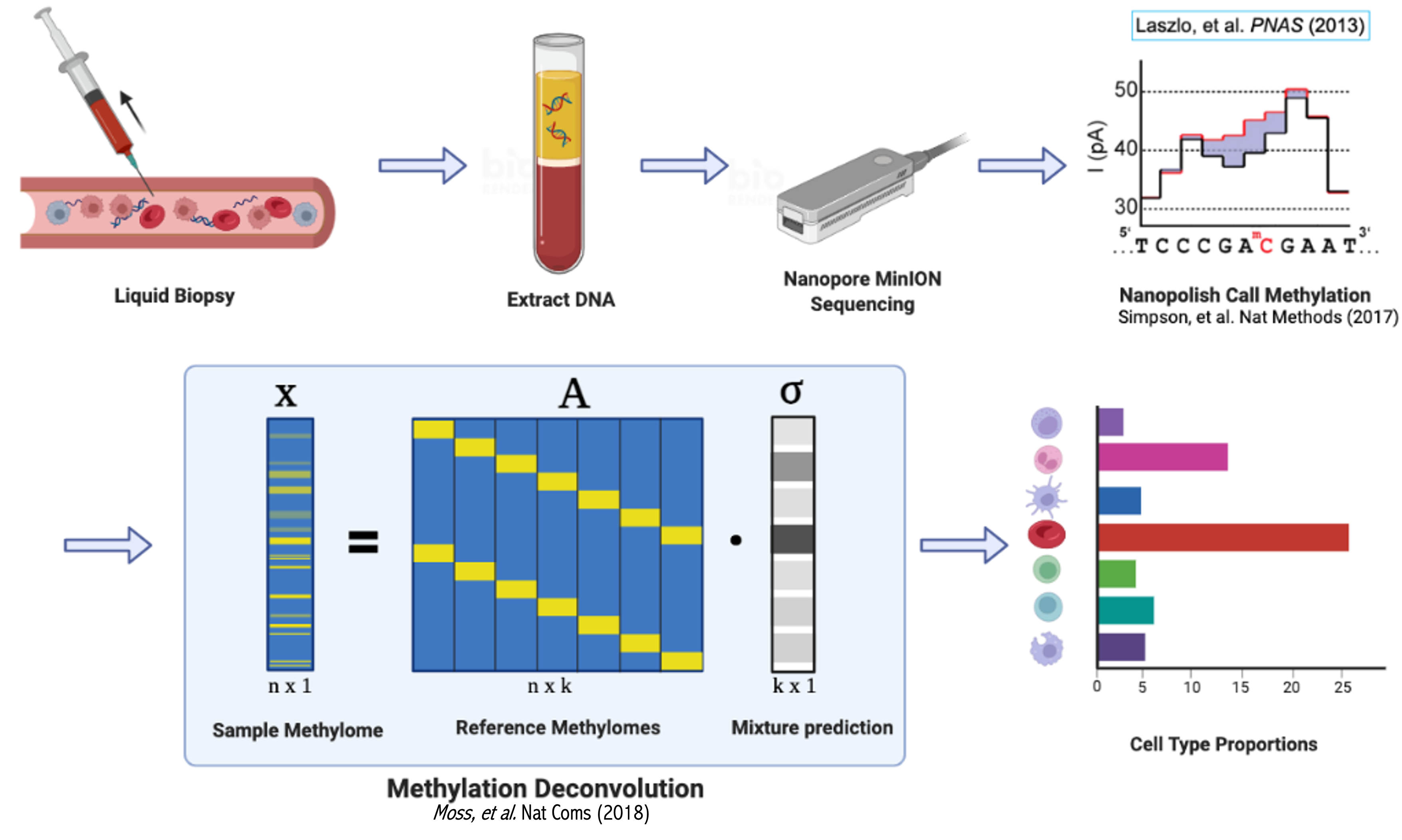
This repository contains a nextflow workflow for deconvoluting a complex mixture of DNA into mixture proportions using nanopore methylation calls. A task useful for cell-free DNA analysis. Input files can either start from raw .fast5 files or basecalled .bam files.
The following programs should be installed into your environment
Make a new directory for your run. Inside that directory make another directory called data:
mkdir dev1
cd dev1
mkdir data
Then inside the data directory make symbolic links to the directories where the nanopore runs are kept.
cd data
ln -s <PATH_TO_DATA>
Then whilst in the run directory execute the nextflow script with the following command which takes as input the path to this repository.
cd ..
nextflow run <.../nanopore_cfdna/>
To view parameters for deconvolution run
nextflow run <.../nanopore_cfdna/> --help
bamsdirectory contains modified bam filesmethylomesdirectory contains the methylome files (counts of modification calls)mixture_proportionscontains sigma vectors for each sampleplotscontains a visualization of the results