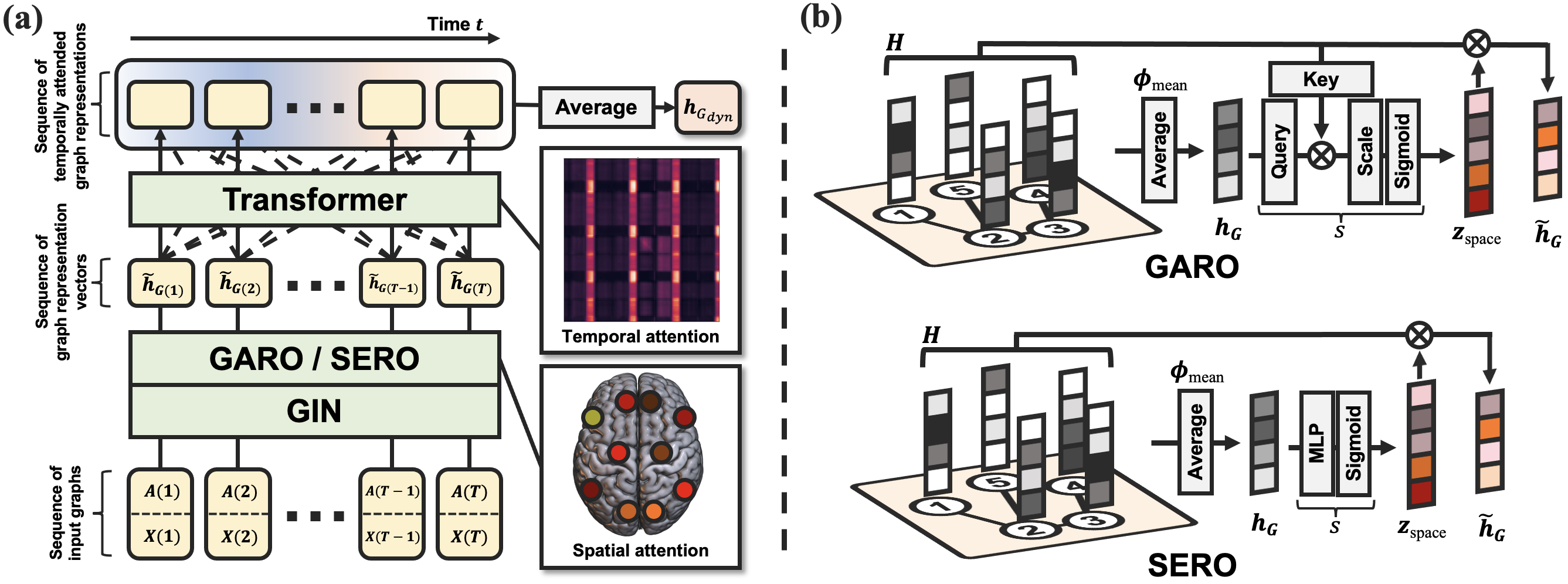
Learning Dynamic Graph Representation of Brain Connectome with Spatio-Temporal Attention
Byung-Hoon Kim, Jong Chul Ye, Jae-Jin Kim
presented at NeurIPS 2021
arXiv, OpenReview, proceeding
The fMRI data used for the experiments of the paper should be downloaded from the Human Connectome Project.
data (specified by option --sourcedir)
├─── behavioral
│ ├─── hcp.csv
│ ├─── hcp_taskrest_EMOTION.csv
│ ├─── hcp_taskrest_GAMBLING.csv
│ ├─── ...
│ └─── hcp_taskrest_WM.csv
├─── img
│ ├─── REST
│ │ ├─── 123456.nii.gz
│ │ ├─── 234567.nii.gz
│ │ ├─── ...
│ │ └─── 999999.nii.gz
│ └─── TASK
│ ├─── EMOTION
│ │ ├─── 123456.nii.gz
│ │ ├─── 234567.nii.gz
│ │ ├─── ...
│ │ └─── 999999.nii.gz
│ ├─── GAMBLING
│ │ ├─── ...
│ │ └─── 999999.nii.gz
│ ├─── ...
│ └─── WM
│ ├─── ...
│ └─── 999999.nii.gz
└───roi
└─── 7_400_coord.csv
| Subject | Gender |
|---------|--------|
| 123456 | F |
| 234567 | M |
| ...... | ...... |
| 999999 | F |
| Task | Rest |
|------|------|
| 0 | 1 |
| 0 | 1 |
| ... | ... |
| 1 | 0 |
| ROI Index | Label Name | R | A | S |
|-----------|----------------------------|---|---|---|
| 0 | NONE | NA| NA| NA|
| 1 | 7Networks_LH_Vis_1 |-32|-42|-20|
| 2 | 7Networks_LH_Vis_2 |-30|-32|-18|
| ... | ......... | . | . | . |
| 400 | 7Networks_RH_Default_PCC_9 | 8 |-50| 44|
Run the main script to perform experiments
python main.pyCommand-line options can be listed with -h flag.
python main.py -h- python 3.8.5
- numpy == 1.20.2
- torch == 1.7.0
- torchvision == 0.8.1
- einops == 0.3.0
- sklearn == 0.24.2
- nilearn == 0.7.1
- nipy == 0.5.0
- pingouin == 0.3.11
- tensorboard == 2.5.0
- tqdm == 4.60.0
For brainplot:
- MRIcroGL >= 1.2
- opencv-python == 4.5.2
- 2022-04-29
5c262d8d: Top k-percentile values from the adjacency matrix is now calculated without the need for calling .detach().cpu().numpy() which improves computation speed.