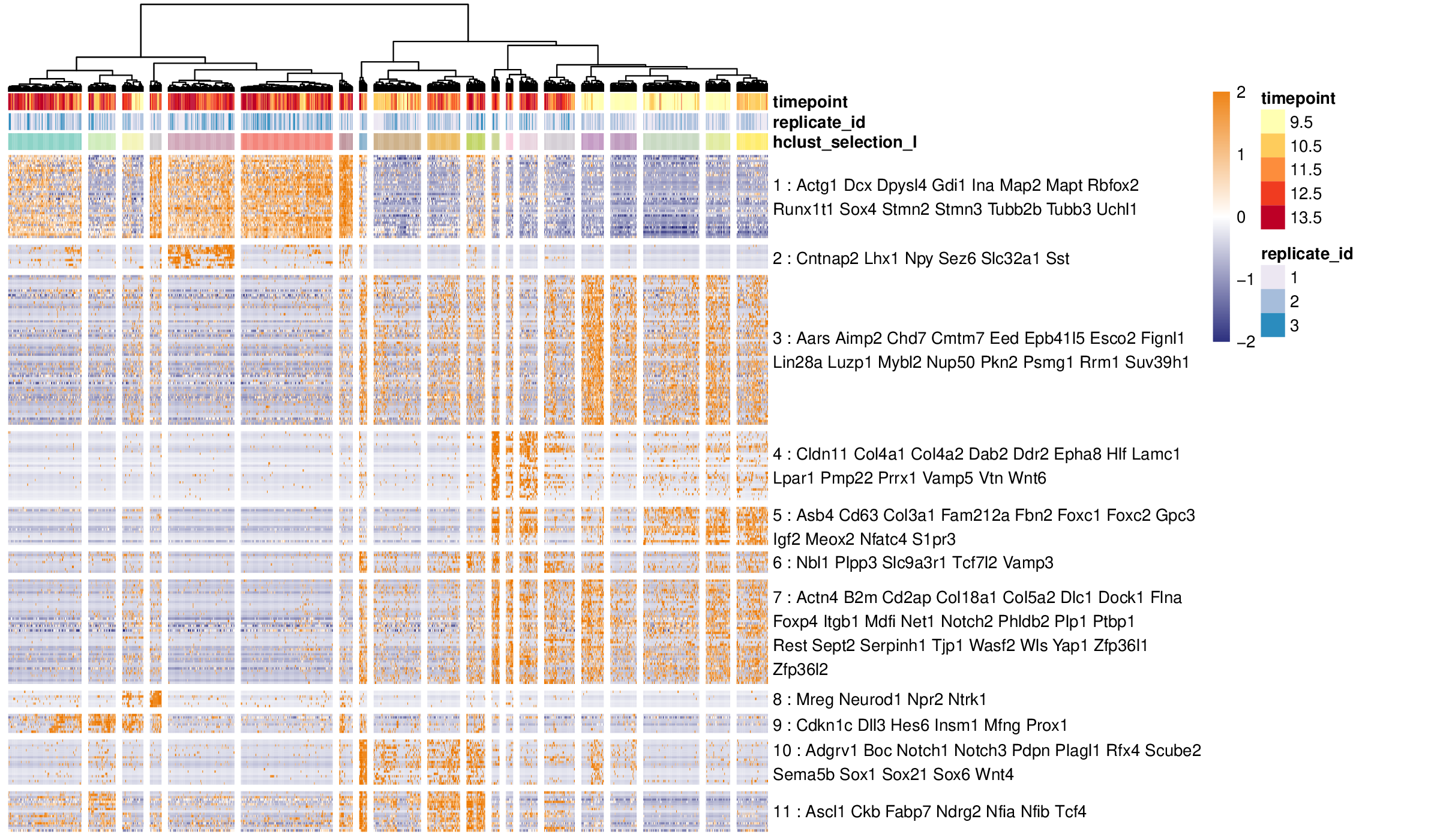
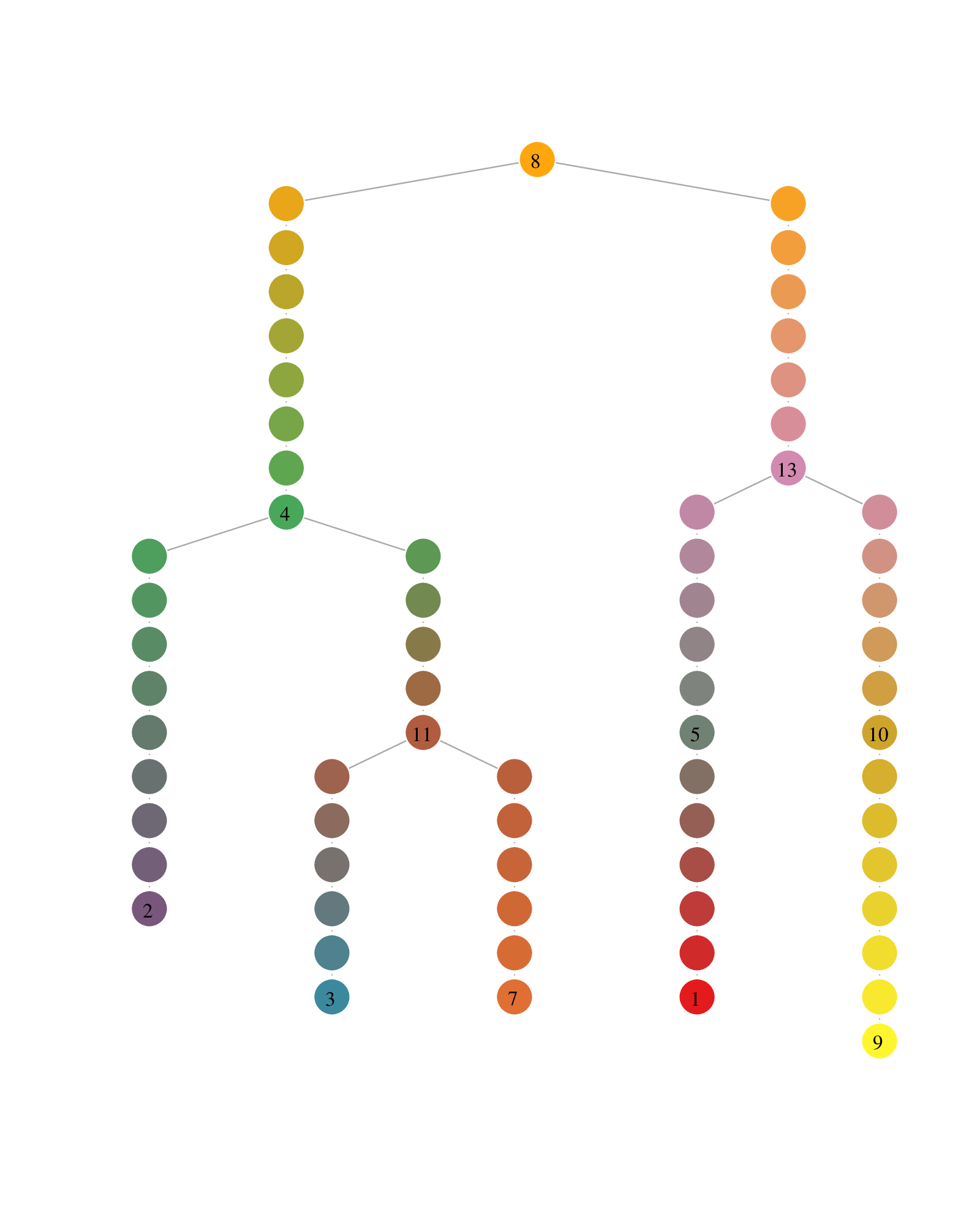
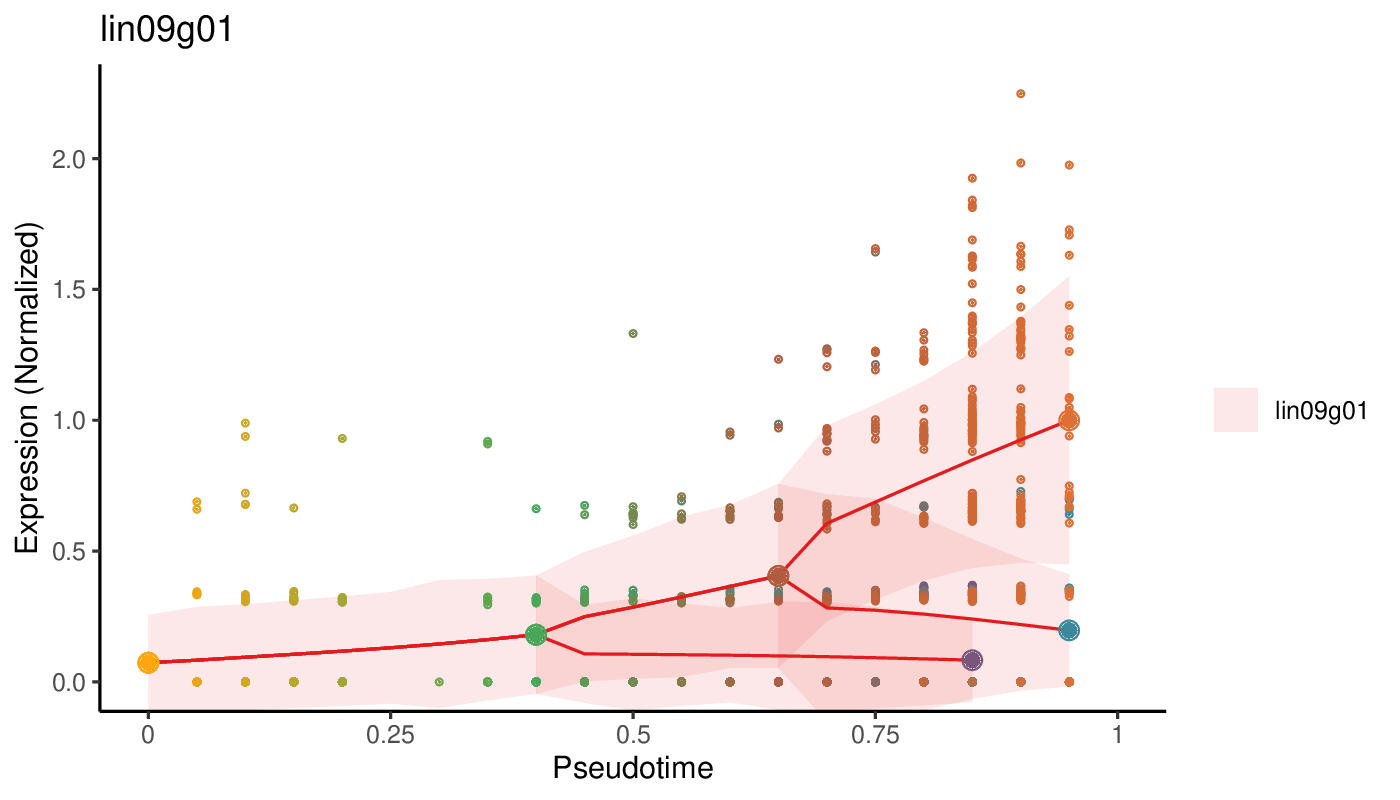
Antler (Another transcriptome lineage explorer) is an R package providing a set of methods for analysing single-cell RNA-seq experiments.
The idea behind Antler is to perform completely unbiased and data-driven analysis to enable the discovery of novel candidate genes and transcriptomic states.
Two vignettes demonstrate how Antler can be used to:
-
Identify, visualize and export the transcriptomic states of a dataset (Link).
-
Build the lineage tree of a cell population differentiating over time and infer the pseudotime dynamics of gene expression (Link).
Antler can be installed from Github via the devtools package:
if (!require("devtools")) {
install.packages("devtools")
}
devtools::install_github("juliendelile/Antler", dependencies = TRUE)Unfortunately, two dependencies (ArrayBin and heatmap.plus) are now only accessible on CRAN archive so they must be installed manually first.
- ArrayBin
# ArrayBin requires SAGx from Bioconductor
install.packages("BiocManager")
BiocManager::install(version = "3.13")
BiocManager::install("SAGx")
devtools::install_version('ArrayBin', version = '0.2')- heatmap.plus
devtools::install_version('heatmap.plus', version = '1.3')Some of the plots produced in the two vignettes include:
- the transcriptomic summary of the cell states
- the cell state graph enabling the pseudotime ordering of the cells.
- the lineage tree
- the reconstructed pseudotime dynamics of a gene