PoreBlazer (v4.0) source code, examples, and geometric properties of porous materials calculated for the subset of 12,000 structures from the CSD MOF.
- Folder: src
Folder containing the complete source code, precompiled executables and associated README_PB_v4.0.txt file with the instructions on how to compile and use the code (see also "How to" below).
- Folder: data
Files containing databases of MOFs and their properties, MOFsubsetPB4.dat, MOFsubsetZeo++.dat, MOFsubsetRASPA.dat
-
Zip file containing complete setup examples for three case study materials HKUST-1, IRMOF-1, ZIF-8: PB4_vs_Zeo++_vs_RASPA.zip
-
Zip file containing a larger collection of case studies (MOFS, zeolites): case_studies.zip
To download the distribution, clone the PoreBlazer repository:
git clone https://github.com/SarkisovGroup/PoreBlazer
cd PoreBlazer
cd srcExecutable poreblazer_gfortran.exe was compiled on Intel Xeon E5-2630 v3 @2.40Hz processor, using gfortran compiler GNU Fortran (GCC) 4.8.5. The poreblazer_intel.exe was compiled using 17.0.4 version of Intel Fortran compiler. These executables should be compatible with most Intel platforms.
If you wish to compile your own version go to the next step.
2.1 Compiler. The Makefile is also provided. To change compiler from gfortran to intel, in the line
FORTRAN_COMPILER= gfortranchange gfortran to the intel compiler executable name on your system.
2.2 Make.To compile the code you can simply issue a "make" command, which will follow the instructions in the Makefile to assemble the code into the poreblazer.exe executable file.
2.3 Other compilers. The code has been tested with Intel fortran compiler and gfortran. Compilation and testing using other compilers is at the discretion of the users. The mutual file dependency is summarized at the end of the Makefile for custom compilations.
The distribution also contains the following case studies (case_studies.zip):
MOFs: CD121, HKUST1, IRMOF1, MOF180, MIL47V, MIL101, ZIF8
Zeolites: BHP, CLO, LOV, ROG, RON, STO, WEI
Other: Slit
with reference results and performance.
2.3 How to run
2.3.1 Basic mode
In basic mode the program uses default parameter setting. For this, in the location of the run, you need to put files defaults.dat and UFF.atoms. They can be copied from the case studies provided with the distribution and are the same for all runs. All that a user needs to specify is the name of the file containing the xyz coordinates of the structure (which uses the standard names for the elements, H, C, O etc) and dimensions of the unit cell (side lengths and yz, xz, xy angles. For example, for HKUST1, it is specified in the input.dat file:
HKUST1.xyz
26.28791 26.28791 26.28791
90 90 90To run simply issue:
./poreblazer.exe < input.datExamples of the output are summarized in the provided case studies.
2.3.2 Advanced mode
In the advanced mode, a user controls various parameters of the simulation, such as the interaction parameters of Nitrogen and Helium atoms, cut-off radius, grid size etc. All the parameters are specified in the defaults.dat file and can be changed there. This file also specifies a source of the force field parameters for the atoms of the structure. By default it is the UFF forcefield, provided in UFF.atoms file (note that not all possible atoms from the UFF are there, but the most common ones). Other force fields and files can be used, with the customized convention for the atom names defined by the user.
This mode assumes the user knows what s/he does. Here is an example of the defaults.dat file:
UFF.atoms Default forcefield: UFF
2.58, 10.22, 298, 12.8 Parameters for the helium atom probe s (Å) and e(K), temperature T (K), required for the Helium porosimetry, and cut-off distance (Å), required for the Helum porosimetry
3.314 Nitrogen atom probe s (Å)
500 Number of trials per atom for the surface area calculation
0.2 Cubelet size
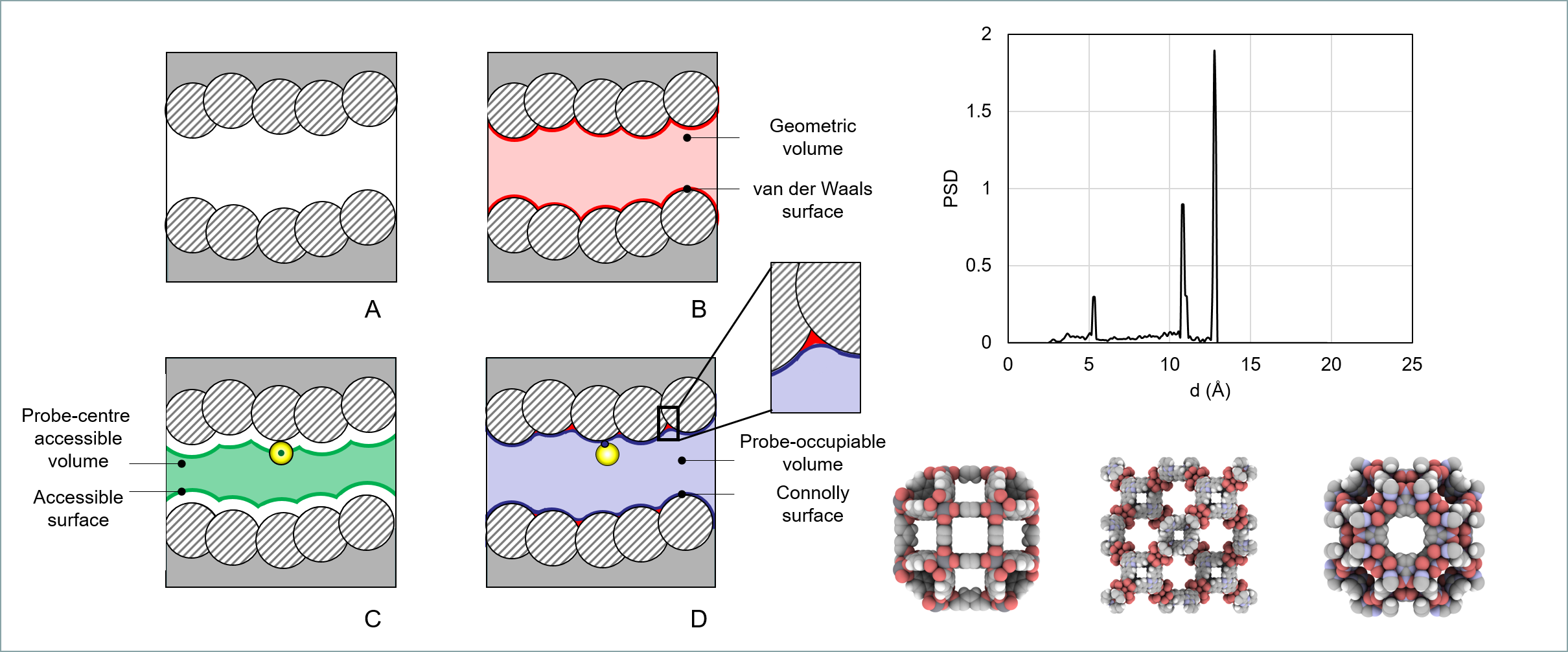
20.0, 0.25 Largest anticipated pore diameter (Å), size of the bin for PSD (Å)
21908391 Rand number generator seed
0 Network-accessible nitrogen probe-centre visualization options: 0 – no network visualization files; 1 – xyz format; 2- grd format; 3 – both xyz and grd format2.3.3 Understanding and interpreting the results
Results are printed on the screen, and can be redirected to a text file. They are also summarized in summary.dat file. Here is an example from HKUST1 with an explanation on what is what:
HKUST1.xyz Name of the structure, file
V_A^3 18166.371 Volume of the system, V, Å
M _g/mol 9674.208 Molecular weight of the unit cell, g/mol
RHO_g/cm^3 0.884 Density, g/cm3
PLD_A 6.38 Pore limiting diameter, Å
LCD_A 12.86 Largest cavity dimeter, Å
D 3 Number of directions the system is percolated in
Total The next properties are total properties
S_AC_A^2 2989.37 Accessible surface area, S_(AC,T), Å2
S_AC_ m^2/cm^3 1645.55 Accessible surface area, S_(AC,T), m2/cm3
S_AC_m^2/g 1860.87 Accessible surface area, S_(AC,T), m2/g
Total The next properties are total properties
V_He_A^3 13419.839 Helium pore volume, V_(He,T), Å3
V_He_cm^3/g 0.835 Helium pore volume, V_(He,T), cm3/g
V_G_A^3 12757.117 Geometric pore volume, V_(G,T), Å3
V_G cm^3/g: 0.794 Geometric pore volume, V_(G,T), cm3/g
V_PO A^3: 12243.005 Probe-occupiable pore volume, V_(PO,T), Å3
V_PO cm^3/g: 0.762 Probe-occupiable volume, V_(PO,T), cm3/g
FV_PO: 0.67394 Volume fraction, , V_(PO,T)/V
Network-accessible The next properties are network-accessible properties
S_AC_A^2 2904.59 Accessible surface area, S_(AC,A), Å2
S_AC_m^2/cm^3 1598.88 Accessible surface area, S_(AC,A), m2/cm3
S_AC_m^2/g 1808.09 Accessible surface area, S_(AC,A), m2/g
Network-accessible The next properties are network-accessible properties
V_He_A^3 13419.839 Helium pore volume, V_(He,A), Å3
V_He_cm^3/g 0.835 Helium pore volume, V_(He,A), cm3/g
V_G_A^3 12755.759 Geometric pore volume, V_(G,A), Å3
V_G cm^3/g: 0.794 Geometric pore volume, V_(G,A), cm3/g
V_PO A^3: 12186.852 Probe-occupiable pore volume, V_(PO,A), Å3
V_PO cm^3/g: 0.759 Probe-occupiable volume, V_(PO,A), cm3/g
FV_PO: 0.67085 Volume fraction, V_(PO,A)/VAdditionally, the code will generate the following files:
- Total_psd_cumulative.txt Total cumulative Pore Size Distribution
- Total_psd.txt Total pore size distribution
- Network-accessible_psd_cumulative.txt Network-accessible cumulative PSD
- Network-accessible_psd.txt Network-accessible PSD
- probe_occupialbe_volume.xyz Points sampled within the network-accessible probe-occupiable volume
- nitrogen_network.xyz Network-accessible nitrogen probe-centre visualization in xyz format (requires correct option in defaults.dat)
- nitrogen_network.grd Network-accessible nitrogen probe-centre visualization in grd format (requires correct option in defaults.dat)
When contributing to this repository, please first discuss the change you wish to make via issue, email, or any other method with the owners of this repository before making a change.
Email:
Lev Sarkisov
Address:
The Department of Chemical Engineering and Analytical Science
The University of Manchester
Sackville Street
Manchester, M13 9PL
This project is licensed under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or at your option) any later version.