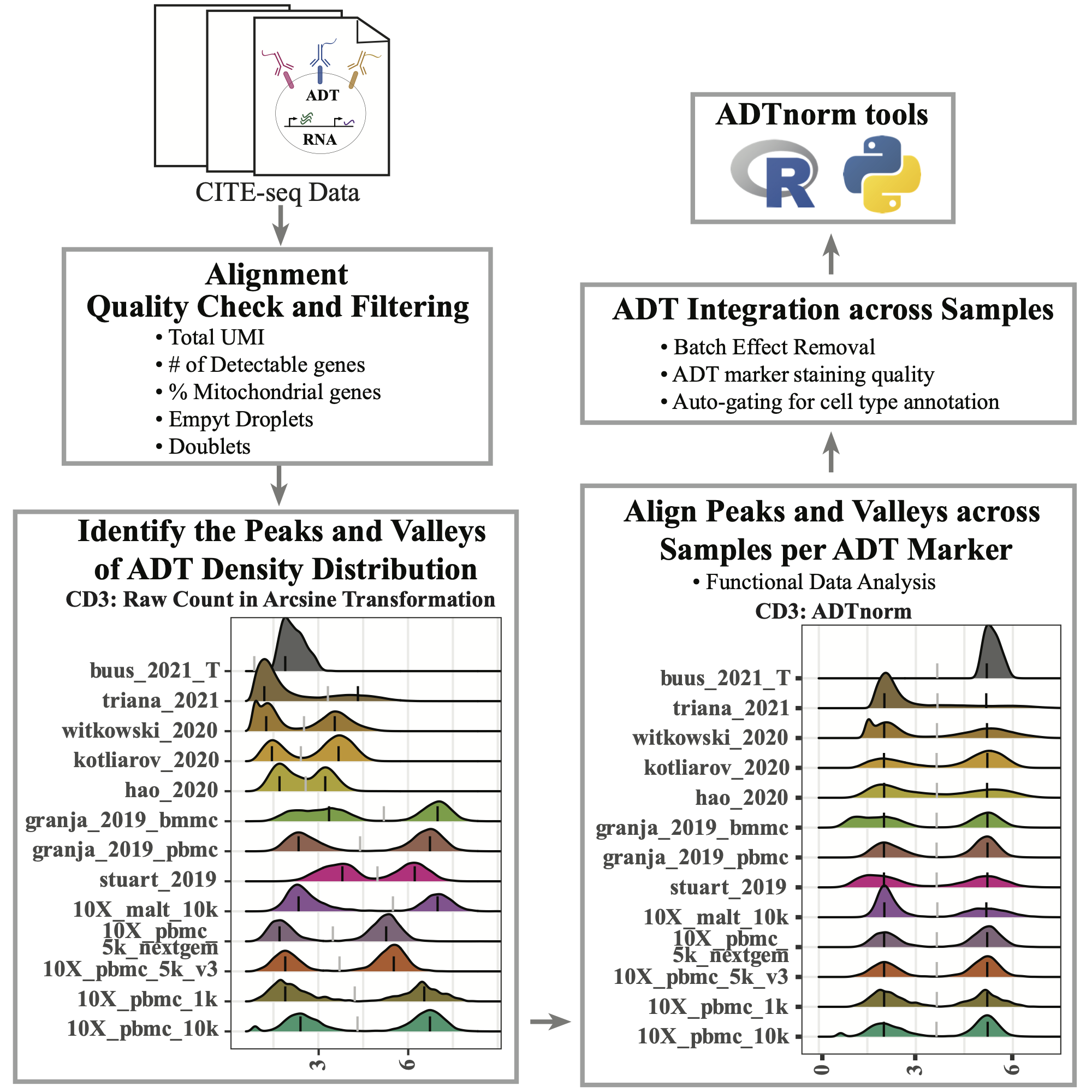
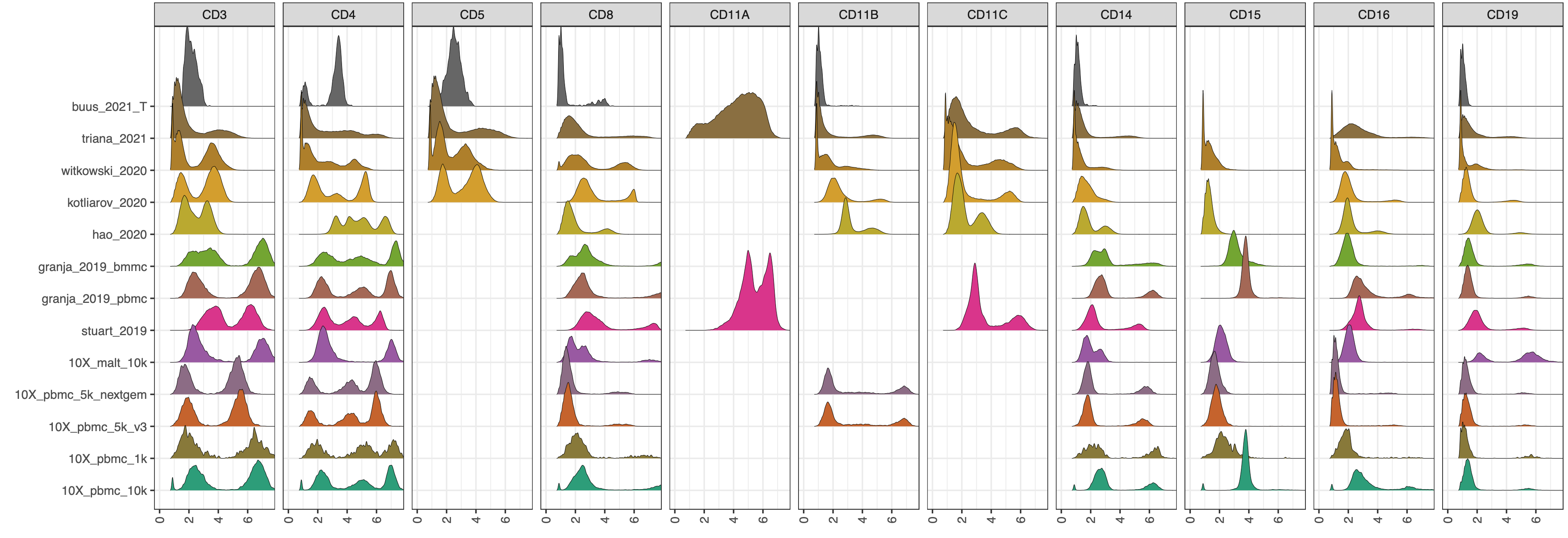
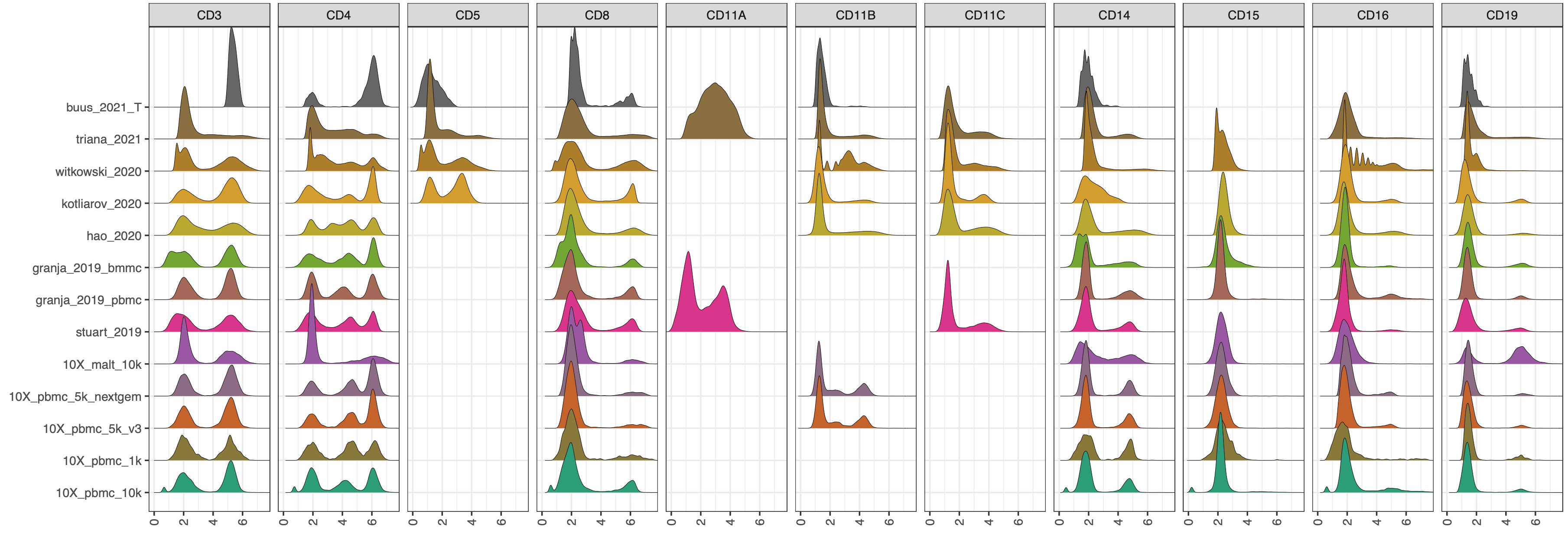
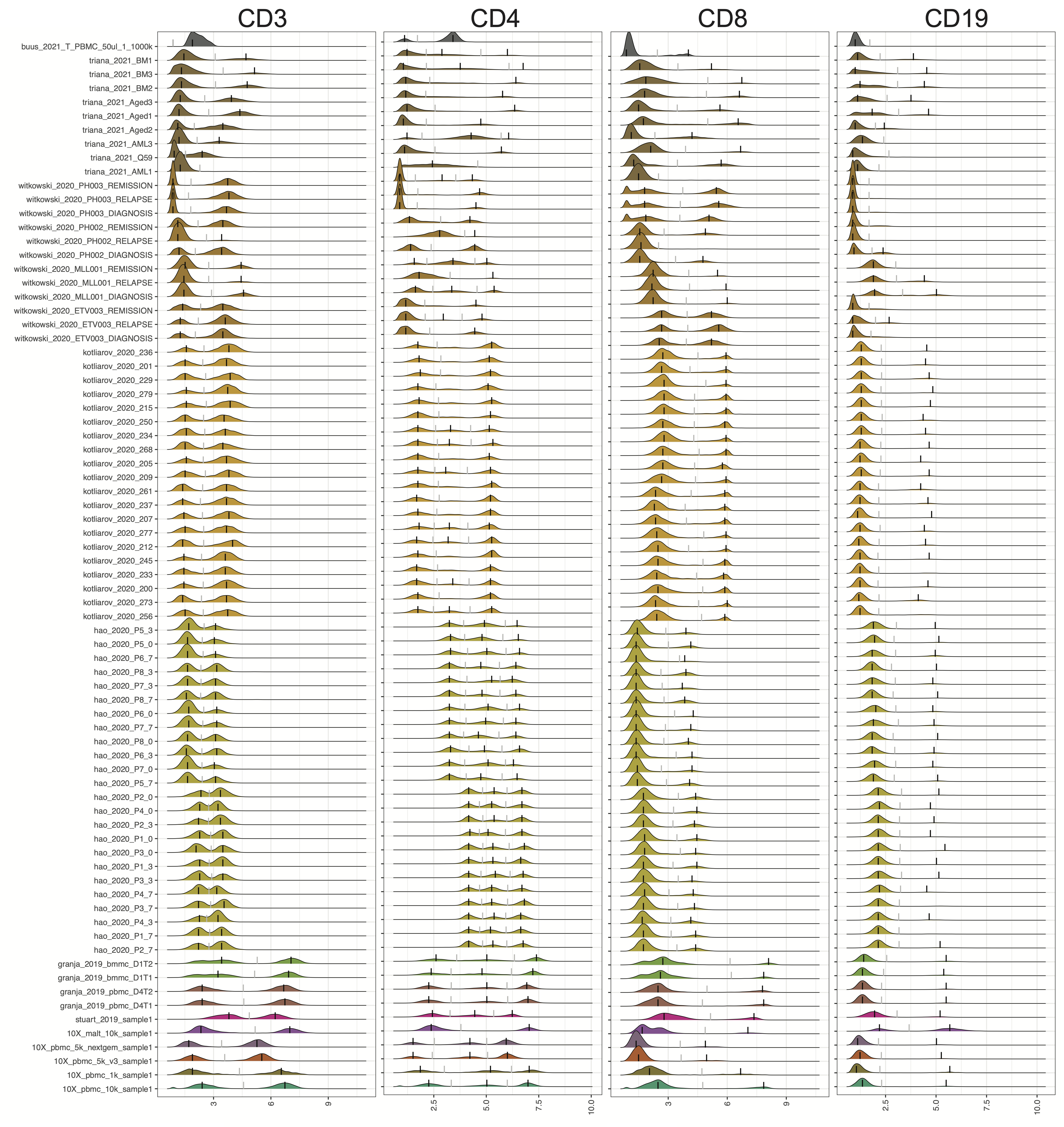
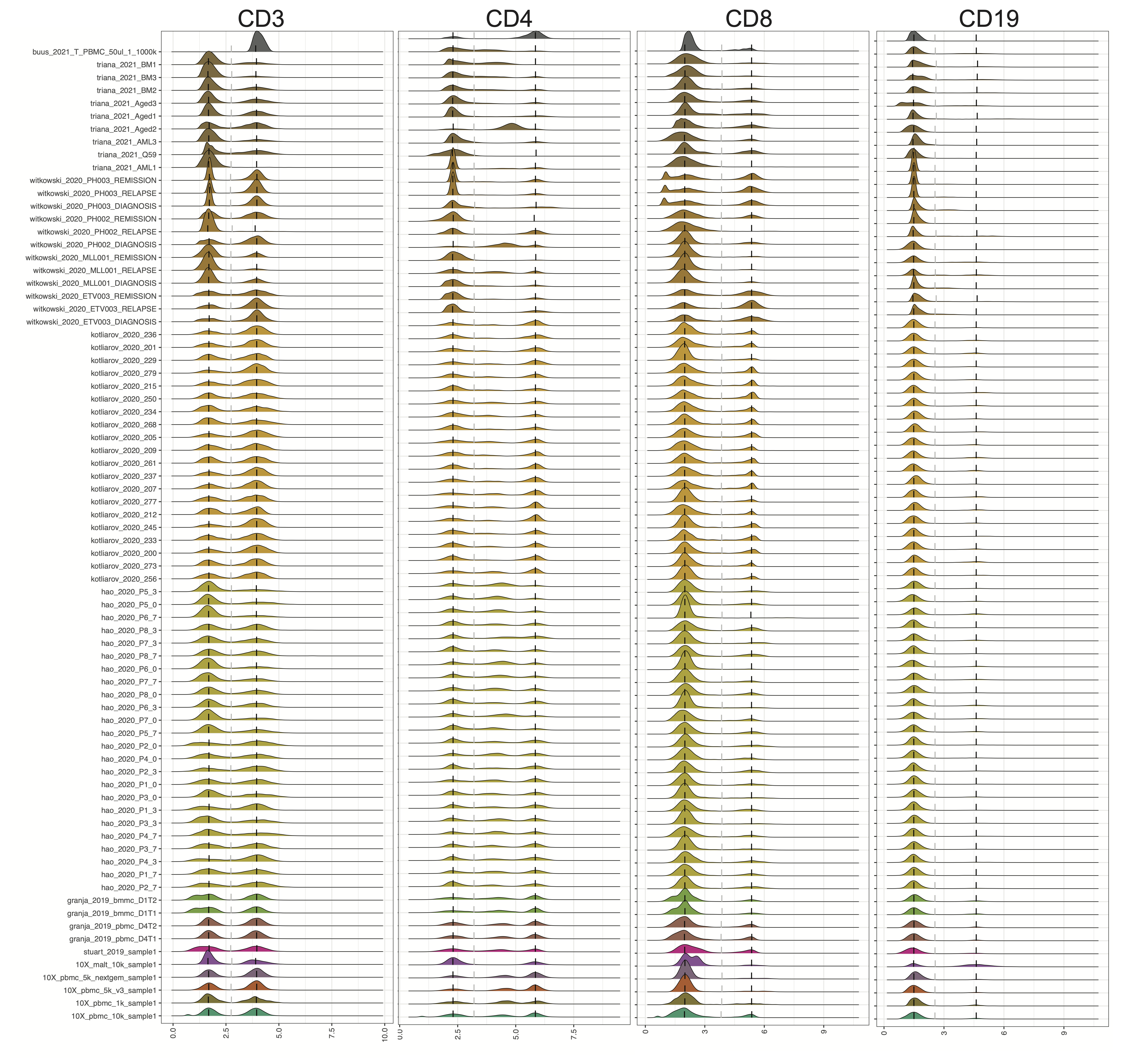
CITE-seq technology enables the direct measurement of protein expression, known as antibody-derived tags (ADT), in addition to RNA expression. The increase in the copy number of protein molecules leads to a more robust detection of protein features compared to RNA, providing a deep definition of cell types. However, due to added discrepancies of antibodies, such as the different types or concentrations of IgG antibodies, the batch effects of the ADT component of CITE-seq can dominate over biological variations, especially for the across-study integration. We present ADTnorm as a normalization and integration method designed explicitly for the ADT counts of CITE-seq data. Benchmarking with existing scaling and normalization methods, ADTnorm achieves a fast and accurate matching of the negative and positive peaks of the ADT counts across samples, efficiently removing technical variations across batches. Further quantitative evaluations confirm that ADTnorm achieves the best cell-type separation while maintaining the minimal batch effect. Therefore, ADTnorm facilitates the scalable ADT count integration of massive public CITE-seq datasets with distinguished experimental designs, which are essential for creating a corpus of well-annotated single-cell data with deep and standardized annotations.
# install.packages("remotes")
remotes::install_github("yezhengSTAT/ADTnorm", build_vignettes = FALSE)The 13 public datasets used in the manuscript is also included in the R package as demo data set. They can be loaded by
data(cell_x_adt)
data(cell_x_feature)
cell_x_adtcontains a matrix of raw count for the cell by ADT markers, which is a data frame with 422682 cells (row) and 9 ADT markers (column): CD3, CD4, CD8, CD14, CD19, CD25, CD45RA, CD56, CD127.
CD3 CD4 CD8 CD14 CD19 CD25 CD45RA CD56 CD127
1 18 138 13 491 3 9 110 17 7
2 30 119 19 472 3 5 125 248 8
3 18 207 10 1289 8 15 5268 26 12
4 18 11 17 20 5 15 4743 491 16
5 5 14 14 19 4 16 4108 458 17
6 21 1014 29 2428 7 52 227 29 15
-
cell_x_featureis a data frame with 422682 cells (row) and 7 feature variables (column):-
sample: Sample name used in original data of each study.
-
batch: Batch information provided from each study.
-
sample_status: Sample status, i.e., Healthy, MALTtumor, HIV Vaccine, Lupus, B-ALL, AML.
-
study_name: Name of the data set/study.
-
ADTseqDepth: Total UMI per cell.
-
cell_type_l1: Broad level of cell type annotation using manual gating.
-
cell_type_l2: Fine level of cell type annotation using manual gating.
-
sample batch sample_status study_name
1 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
2 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
3 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
4 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
5 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
6 10X_pbmc_10k_sample1 10X_pbmc_10k_batch1 healthy 10X_pbmc_10k
ADTseqDepth cell_type_l1 cell_type_l2
1 981 monocytes classical monocyte
2 1475 monocytes classical monocyte
3 7149 monocytes classical monocyte
4 6831 NK CD16+ NK
5 6839 NK CD16+ NK
6 4720 monocytes classical monocyte
library(ADTnorm)
save_outpath <- "/path/to/output/location"
run_name <- "ADTnorm_demoRun"
data(cell_x_adt)
data(cell_x_feature)
cell_x_feature$sample = factor(cell_x_feature$study_name)
cell_x_feature$batch = factor(cell_x_feature$study_name)
cell_x_adt_norm <- ADTnorm(
cell_x_adt = cell_x_adt,
cell_x_feature = cell_x_feature,
save_outpath = save_outpath,
study_name = run_name,
marker_to_process = c("CD3", "CD4", "CD8", "CD45RA"),
trimodal_marker = c("CD4", "CD45RA"),
positive_peak = list(ADT = "CD3", sample = "buus_2021_T"),
save_intermediate_fig = TRUE
)library(ADTnorm)
save_outpath <- "/path/to/output/location"
run_name <- "ADTnorm_demoRun"
data(cell_x_adt)
data(cell_x_feature)
cell_x_feature$batch = factor(cell_x_feature$study_name)
cell_x_adt_norm <- ADTnorm(
cell_x_adt = cell_x_adt,
cell_x_feature = cell_x_feature,
save_outpath = save_outpath,
study_name = run_name,
marker_to_process = c("CD3", "CD4", "CD8", "CD45RA"),
trimodal_marker = c("CD4", "CD45RA"),
positive_peak = list(ADT = "CD3", sample = "buus_2021_T"),
save_intermediate_fig = TRUE
)Basic parameters introduction. The full parameter explanation for the ADTnorm function can be found at Reference - ADTnorm.
cell_x_adt: Matrix of ADT raw counts in cells (rows) by ADT markers (columns) format.
cell_x_feature: Matrix of cells (rows) by cell features (columns) such as cell type, sample, and batch-related information.
save_outpath: The path to save the results.
study_name: Name of this run.
marker_to_process: Markers to normalize. Leaving empty to process all the ADT markers in cell_x_adt matrix.
bimodal_marker: Specify ADT markers that are likely to have two peaks based on researchers' prior knowledge or preliminary observation of particular data to be processed. Leaving it as default, ADTnorm will try to find the bimodal peak in all markers that are not listed in `trimodal_marker.`
trimodal_marker: Index of the ADT markers that tend to have three peaks based on researchers' prior knowledge (e.g., CD4) or preliminary observation on particular data to be processed.
positive_peak: A list variable containing a vector of ADT marker(s) and a corresponding vector of sample name(s) in matching order to specify that the uni-peak detected should be aligned to positive peaks. For example, for samples that only contain T cells. The only CD3 peak should be aligned to the positive peaks of other samples.
save_intermediate_fig: Save the density plot figure for checking the peak and valley location detection.
For more detailed and typical parameter tuning examples, please visit tutorial website. We will illustrate using the demo data.
ADTnorm function will generate a matrix of rows of the same number as input cell_x_adt row number and columns are ADT markers specificed in marker_to_process. The value in the matrix is normalized value by ADTnorm. In the save_outpath specified by the users, there will be two subfolders, figures and RDS, containing the intermediate object and density plot of detected peak and valley landmarks before and after ADTnorm. Those figures can be used to check if further parameter tuning is needed for certain ADT markers.
Email: yzheng23@fredhutch.org
Twitter: @yezhengSTAT